Model info
| Transcription factor | Batf3 | ||||||||
| Model | BATF3_MOUSE.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 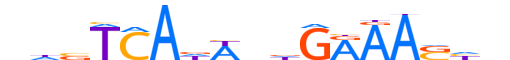 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 17 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndSTTYChnWhTGAbdn | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.963 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 5 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | B-ATF-related factors {1.1.4} | ||||||||
| TF subfamily | B-ATF-3 {1.1.4.0.3} | ||||||||
| MGI | MGI:1925491 | ||||||||
| EntrezGene | GeneID:381319 (SSTAR profile) | ||||||||
| UniProt ID | BATF3_MOUSE | ||||||||
| UniProt AC | Q9D275 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Batf3 expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 86.0 | 177.0 | 103.0 | 134.0 |
| 02 | 242.0 | 28.0 | 122.0 | 108.0 |
| 03 | 15.0 | 195.0 | 251.0 | 39.0 |
| 04 | 15.0 | 8.0 | 7.0 | 470.0 |
| 05 | 23.0 | 32.0 | 4.0 | 441.0 |
| 06 | 21.0 | 80.0 | 21.0 | 378.0 |
| 07 | 9.0 | 428.0 | 10.0 | 53.0 |
| 08 | 251.0 | 89.0 | 60.0 | 100.0 |
| 09 | 105.0 | 91.0 | 109.0 | 195.0 |
| 10 | 115.0 | 21.0 | 32.0 | 332.0 |
| 11 | 241.0 | 99.0 | 28.0 | 132.0 |
| 12 | 4.0 | 0.0 | 5.0 | 491.0 |
| 13 | 6.0 | 12.0 | 405.0 | 77.0 |
| 14 | 432.0 | 3.0 | 21.0 | 44.0 |
| 15 | 50.0 | 231.0 | 159.0 | 60.0 |
| 16 | 126.0 | 60.0 | 73.0 | 241.0 |
| 17 | 121.0 | 193.0 | 93.0 | 93.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.368 | 0.344 | -0.191 | 0.069 |
| 02 | 0.655 | -1.454 | -0.024 | -0.144 |
| 03 | -2.034 | 0.44 | 0.691 | -1.138 |
| 04 | -2.034 | -2.584 | -2.694 | 1.315 |
| 05 | -1.64 | -1.328 | -3.126 | 1.252 |
| 06 | -1.725 | -0.439 | -1.725 | 1.098 |
| 07 | -2.484 | 1.222 | -2.394 | -0.841 |
| 08 | 0.691 | -0.335 | -0.721 | -0.22 |
| 09 | -0.172 | -0.313 | -0.135 | 0.44 |
| 10 | -0.082 | -1.725 | -1.328 | 0.969 |
| 11 | 0.651 | -0.23 | -1.454 | 0.054 |
| 12 | -3.126 | -4.4 | -2.961 | 1.359 |
| 13 | -2.819 | -2.234 | 1.167 | -0.477 |
| 14 | 1.231 | -3.325 | -1.725 | -1.022 |
| 15 | -0.898 | 0.608 | 0.238 | -0.721 |
| 16 | 0.008 | -0.721 | -0.529 | 0.651 |
| 17 | -0.032 | 0.43 | -0.291 | -0.291 |