Model info
| Transcription factor | Clock | ||||||||
| Model | CLOCK_MOUSE.H11MO.0.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 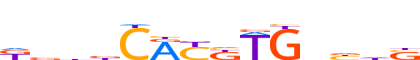 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 14 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | SdvnCAMRYGvvvh | ||||||||
| Best auROC (human) | 0.658 | ||||||||
| Best auROC (mouse) | 0.735 | ||||||||
| Peak sets in benchmark (human) | 5 | ||||||||
| Peak sets in benchmark (mouse) | 22 | ||||||||
| Aligned words | 253 | ||||||||
| TF family | PAS domain factors {1.2.5} | ||||||||
| TF subfamily | Arnt-like factors {1.2.5.2} | ||||||||
| MGI | MGI:99698 | ||||||||
| EntrezGene | GeneID:12753 (SSTAR profile) | ||||||||
| UniProt ID | CLOCK_MOUSE | ||||||||
| UniProt AC | O08785 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Clock expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 40.0 | 151.0 | 45.0 | 16.0 |
| 02 | 107.0 | 29.0 | 48.0 | 68.0 |
| 03 | 51.0 | 37.0 | 129.0 | 35.0 |
| 04 | 85.0 | 81.0 | 42.0 | 44.0 |
| 05 | 24.0 | 214.0 | 11.0 | 3.0 |
| 06 | 204.0 | 16.0 | 11.0 | 21.0 |
| 07 | 39.0 | 153.0 | 32.0 | 28.0 |
| 08 | 64.0 | 14.0 | 162.0 | 12.0 |
| 09 | 24.0 | 30.0 | 11.0 | 187.0 |
| 10 | 22.0 | 6.0 | 217.0 | 7.0 |
| 11 | 101.0 | 79.0 | 45.0 | 27.0 |
| 12 | 93.0 | 61.0 | 71.0 | 27.0 |
| 13 | 56.0 | 98.0 | 73.0 | 25.0 |
| 14 | 124.0 | 75.0 | 6.0 | 47.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.442 | 0.862 | -0.328 | -1.309 |
| 02 | 0.521 | -0.751 | -0.265 | 0.075 |
| 03 | -0.206 | -0.517 | 0.706 | -0.571 |
| 04 | 0.294 | 0.247 | -0.395 | -0.35 |
| 05 | -0.931 | 1.208 | -1.649 | -2.687 |
| 06 | 1.16 | -1.309 | -1.649 | -1.057 |
| 07 | -0.466 | 0.875 | -0.657 | -0.784 |
| 08 | 0.015 | -1.432 | 0.931 | -1.571 |
| 09 | -0.931 | -0.719 | -1.649 | 1.074 |
| 10 | -1.013 | -2.166 | 1.221 | -2.039 |
| 11 | 0.464 | 0.222 | -0.328 | -0.819 |
| 12 | 0.383 | -0.032 | 0.117 | -0.819 |
| 13 | -0.115 | 0.434 | 0.144 | -0.892 |
| 14 | 0.667 | 0.171 | -2.166 | -0.286 |