Model info
| Transcription factor | E2F8 (GeneCards) | ||||||||
| Model | E2F8_HUMAN.H11MO.0.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 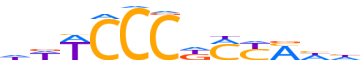 | ||||||||
| Data source | HT-SELEX | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 12 | ||||||||
| Quality | D | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dnKRKMGGGWWd | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 28 | ||||||||
| TF family | E2F-related factors {3.3.2} | ||||||||
| TF subfamily | E2F {3.3.2.1} | ||||||||
| HGNC | HGNC:24727 | ||||||||
| EntrezGene | GeneID:79733 (SSTAR profile) | ||||||||
| UniProt ID | E2F8_HUMAN | ||||||||
| UniProt AC | A0AVK6 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | E2F8 expression | ||||||||
| ReMap ChIP-seq dataset list | E2F8 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 6.5 | 3.5 | 4.5 | 13.5 |
| 02 | 5.0 | 3.0 | 5.0 | 15.0 |
| 03 | 1.25 | 3.25 | 4.25 | 19.25 |
| 04 | 8.5 | 0.5 | 16.5 | 2.5 |
| 05 | 3.0 | 0.0 | 19.0 | 6.0 |
| 06 | 9.0 | 13.0 | 0.0 | 6.0 |
| 07 | 0.0 | 1.0 | 26.0 | 1.0 |
| 08 | 0.0 | 0.0 | 27.0 | 1.0 |
| 09 | 0.0 | 0.0 | 25.0 | 3.0 |
| 10 | 19.25 | 0.25 | 1.25 | 7.25 |
| 11 | 16.25 | 4.25 | 2.25 | 5.25 |
| 12 | 12.25 | 3.25 | 4.25 | 8.25 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.066 | -0.592 | -0.384 | 0.604 |
| 02 | -0.295 | -0.715 | -0.295 | 0.704 |
| 03 | -1.325 | -0.652 | -0.432 | 0.942 |
| 04 | 0.175 | -1.771 | 0.794 | -0.854 |
| 05 | -0.715 | -2.241 | 0.929 | -0.137 |
| 06 | 0.227 | 0.569 | -2.241 | -0.137 |
| 07 | -2.241 | -1.452 | 1.231 | -1.452 |
| 08 | -2.241 | -2.241 | 1.268 | -1.452 |
| 09 | -2.241 | -2.241 | 1.193 | -0.715 |
| 10 | 0.942 | -1.979 | -1.325 | 0.031 |
| 11 | 0.78 | -0.432 | -0.932 | -0.253 |
| 12 | 0.513 | -0.652 | -0.432 | 0.148 |