Model info
| Transcription factor | Egr2 | ||||||||
| Model | EGR2_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 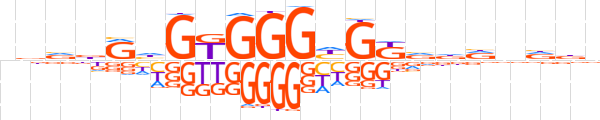 | ||||||||
| LOGO (reverse complement) | 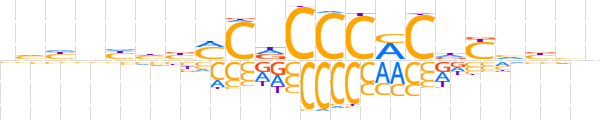 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 21 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vdvdRhGKGGGhGKvddnndv | ||||||||
| Best auROC (human) | 0.961 | ||||||||
| Best auROC (mouse) | 0.996 | ||||||||
| Peak sets in benchmark (human) | 4 | ||||||||
| Peak sets in benchmark (mouse) | 25 | ||||||||
| Aligned words | 575 | ||||||||
| TF family | Three-zinc finger Krüppel-related factors {2.3.1} | ||||||||
| TF subfamily | EGR factors {2.3.1.3} | ||||||||
| MGI | MGI:95296 | ||||||||
| EntrezGene | GeneID:13654 (SSTAR profile) | ||||||||
| UniProt ID | EGR2_MOUSE | ||||||||
| UniProt AC | P08152 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Egr2 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 21.0 | 8.0 | 37.0 | 21.0 | 32.0 | 16.0 | 29.0 | 19.0 | 50.0 | 35.0 | 76.0 | 54.0 | 5.0 | 6.0 | 46.0 | 24.0 |
| 02 | 18.0 | 17.0 | 63.0 | 10.0 | 27.0 | 16.0 | 18.0 | 4.0 | 41.0 | 33.0 | 97.0 | 17.0 | 9.0 | 16.0 | 74.0 | 19.0 |
| 03 | 12.0 | 9.0 | 53.0 | 21.0 | 16.0 | 10.0 | 11.0 | 45.0 | 41.0 | 31.0 | 87.0 | 93.0 | 7.0 | 4.0 | 18.0 | 21.0 |
| 04 | 13.0 | 7.0 | 50.0 | 6.0 | 10.0 | 5.0 | 37.0 | 2.0 | 31.0 | 13.0 | 118.0 | 7.0 | 17.0 | 19.0 | 141.0 | 3.0 |
| 05 | 33.0 | 14.0 | 6.0 | 18.0 | 4.0 | 16.0 | 2.0 | 22.0 | 82.0 | 137.0 | 9.0 | 118.0 | 2.0 | 6.0 | 6.0 | 4.0 |
| 06 | 7.0 | 3.0 | 102.0 | 9.0 | 6.0 | 1.0 | 165.0 | 1.0 | 0.0 | 0.0 | 23.0 | 0.0 | 1.0 | 1.0 | 159.0 | 1.0 |
| 07 | 0.0 | 1.0 | 11.0 | 2.0 | 0.0 | 0.0 | 1.0 | 4.0 | 5.0 | 6.0 | 151.0 | 287.0 | 3.0 | 1.0 | 6.0 | 1.0 |
| 08 | 2.0 | 1.0 | 3.0 | 2.0 | 0.0 | 0.0 | 8.0 | 0.0 | 12.0 | 0.0 | 154.0 | 3.0 | 12.0 | 0.0 | 278.0 | 4.0 |
| 09 | 0.0 | 0.0 | 24.0 | 2.0 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 3.0 | 417.0 | 21.0 | 0.0 | 0.0 | 9.0 | 0.0 |
| 10 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 2.0 | 1.0 | 0.0 | 7.0 | 3.0 | 440.0 | 0.0 | 2.0 | 0.0 | 21.0 | 0.0 |
| 11 | 3.0 | 0.0 | 0.0 | 6.0 | 1.0 | 0.0 | 0.0 | 4.0 | 105.0 | 194.0 | 4.0 | 162.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 12 | 5.0 | 4.0 | 95.0 | 5.0 | 9.0 | 5.0 | 170.0 | 10.0 | 0.0 | 1.0 | 2.0 | 1.0 | 1.0 | 4.0 | 163.0 | 4.0 |
| 13 | 0.0 | 0.0 | 10.0 | 5.0 | 3.0 | 1.0 | 4.0 | 6.0 | 29.0 | 13.0 | 278.0 | 110.0 | 4.0 | 2.0 | 11.0 | 3.0 |
| 14 | 8.0 | 6.0 | 17.0 | 5.0 | 4.0 | 3.0 | 6.0 | 3.0 | 91.0 | 39.0 | 147.0 | 26.0 | 13.0 | 16.0 | 73.0 | 22.0 |
| 15 | 28.0 | 6.0 | 70.0 | 12.0 | 18.0 | 14.0 | 12.0 | 20.0 | 31.0 | 24.0 | 132.0 | 56.0 | 6.0 | 12.0 | 22.0 | 16.0 |
| 16 | 12.0 | 8.0 | 49.0 | 14.0 | 14.0 | 7.0 | 25.0 | 10.0 | 41.0 | 33.0 | 133.0 | 29.0 | 8.0 | 12.0 | 69.0 | 15.0 |
| 17 | 18.0 | 9.0 | 33.0 | 15.0 | 15.0 | 18.0 | 10.0 | 17.0 | 74.0 | 49.0 | 80.0 | 73.0 | 11.0 | 10.0 | 35.0 | 12.0 |
| 18 | 21.0 | 18.0 | 68.0 | 11.0 | 14.0 | 16.0 | 39.0 | 17.0 | 36.0 | 18.0 | 87.0 | 17.0 | 9.0 | 14.0 | 73.0 | 21.0 |
| 19 | 18.0 | 6.0 | 45.0 | 11.0 | 9.0 | 15.0 | 18.0 | 24.0 | 40.0 | 37.0 | 139.0 | 51.0 | 8.0 | 9.0 | 35.0 | 14.0 |
| 20 | 16.0 | 11.0 | 30.0 | 18.0 | 11.0 | 19.0 | 28.0 | 9.0 | 55.0 | 57.0 | 92.0 | 33.0 | 6.0 | 20.0 | 54.0 | 20.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.349 | -1.285 | 0.209 | -0.349 | 0.066 | -0.616 | -0.031 | -0.447 | 0.508 | 0.154 | 0.924 | 0.584 | -1.728 | -1.558 | 0.425 | -0.218 |
| 02 | -0.5 | -0.556 | 0.737 | -1.071 | -0.102 | -0.616 | -0.5 | -1.934 | 0.311 | 0.096 | 1.167 | -0.556 | -1.173 | -0.616 | 0.897 | -0.447 |
| 03 | -0.895 | -1.173 | 0.566 | -0.349 | -0.616 | -1.071 | -0.98 | 0.403 | 0.311 | 0.034 | 1.058 | 1.125 | -1.412 | -1.934 | -0.5 | -0.349 |
| 04 | -0.818 | -1.412 | 0.508 | -1.558 | -1.071 | -1.728 | 0.209 | -2.542 | 0.034 | -0.818 | 1.362 | -1.412 | -0.556 | -0.447 | 1.54 | -2.192 |
| 05 | 0.096 | -0.746 | -1.558 | -0.5 | -1.934 | -0.616 | -2.542 | -0.303 | 0.999 | 1.511 | -1.173 | 1.362 | -2.542 | -1.558 | -1.558 | -1.934 |
| 06 | -1.412 | -2.192 | 1.217 | -1.173 | -1.558 | -3.086 | 1.696 | -3.086 | -4.365 | -4.365 | -0.26 | -4.365 | -3.086 | -3.086 | 1.659 | -3.086 |
| 07 | -4.365 | -3.086 | -0.98 | -2.542 | -4.365 | -4.365 | -3.086 | -1.934 | -1.728 | -1.558 | 1.608 | 2.249 | -2.192 | -3.086 | -1.558 | -3.086 |
| 08 | -2.542 | -3.086 | -2.192 | -2.542 | -4.365 | -4.365 | -1.285 | -4.365 | -0.895 | -4.365 | 1.628 | -2.192 | -0.895 | -4.365 | 2.217 | -1.934 |
| 09 | -4.365 | -4.365 | -0.218 | -2.542 | -3.086 | -4.365 | -4.365 | -4.365 | -2.542 | -2.192 | 2.622 | -0.349 | -4.365 | -4.365 | -1.173 | -4.365 |
| 10 | -4.365 | -4.365 | -2.192 | -4.365 | -4.365 | -2.542 | -3.086 | -4.365 | -1.412 | -2.192 | 2.676 | -4.365 | -2.542 | -4.365 | -0.349 | -4.365 |
| 11 | -2.192 | -4.365 | -4.365 | -1.558 | -3.086 | -4.365 | -4.365 | -1.934 | 1.246 | 1.858 | -1.934 | 1.678 | -4.365 | -4.365 | -4.365 | -4.365 |
| 12 | -1.728 | -1.934 | 1.146 | -1.728 | -1.173 | -1.728 | 1.726 | -1.071 | -4.365 | -3.086 | -2.542 | -3.086 | -3.086 | -1.934 | 1.684 | -1.934 |
| 13 | -4.365 | -4.365 | -1.071 | -1.728 | -2.192 | -3.086 | -1.934 | -1.558 | -0.031 | -0.818 | 2.217 | 1.292 | -1.934 | -2.542 | -0.98 | -2.192 |
| 14 | -1.285 | -1.558 | -0.556 | -1.728 | -1.934 | -2.192 | -1.558 | -2.192 | 1.103 | 0.261 | 1.581 | -0.139 | -0.818 | -0.616 | 0.884 | -0.303 |
| 15 | -0.066 | -1.558 | 0.842 | -0.895 | -0.5 | -0.746 | -0.895 | -0.397 | 0.034 | -0.218 | 1.474 | 0.62 | -1.558 | -0.895 | -0.303 | -0.616 |
| 16 | -0.895 | -1.285 | 0.488 | -0.746 | -0.746 | -1.412 | -0.178 | -1.071 | 0.311 | 0.096 | 1.481 | -0.031 | -1.285 | -0.895 | 0.828 | -0.678 |
| 17 | -0.5 | -1.173 | 0.096 | -0.678 | -0.678 | -0.5 | -1.071 | -0.556 | 0.897 | 0.488 | 0.975 | 0.884 | -0.98 | -1.071 | 0.154 | -0.895 |
| 18 | -0.349 | -0.5 | 0.813 | -0.98 | -0.746 | -0.616 | 0.261 | -0.556 | 0.182 | -0.5 | 1.058 | -0.556 | -1.173 | -0.746 | 0.884 | -0.349 |
| 19 | -0.5 | -1.558 | 0.403 | -0.98 | -1.173 | -0.678 | -0.5 | -0.218 | 0.287 | 0.209 | 1.525 | 0.527 | -1.285 | -1.173 | 0.154 | -0.746 |
| 20 | -0.616 | -0.98 | 0.002 | -0.5 | -0.98 | -0.447 | -0.066 | -1.173 | 0.602 | 0.638 | 1.114 | 0.096 | -1.558 | -0.397 | 0.584 | -0.397 |