Retracted!
Do not use this motif!
Do not use this motif!
Model info
| Transcription factor | EVX2 (GeneCards) | ||||||||
| Model | EVX2_HUMAN.H11DI.0.A RETRACTED!!! | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 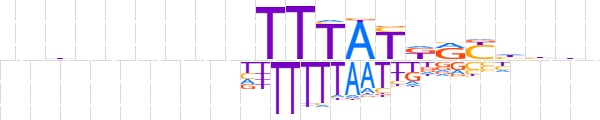 | ||||||||
| LOGO (reverse complement) | 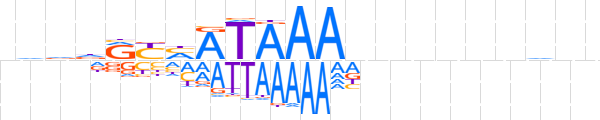 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 21 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nndnnnnnnTTTATKRYhbhn | ||||||||
| Best auROC (human) | 0.974 | ||||||||
| Best auROC (mouse) | 0.883 | ||||||||
| Peak sets in benchmark (human) | 11 | ||||||||
| Peak sets in benchmark (mouse) | 4 | ||||||||
| Aligned words | 494 | ||||||||
| TF family | HOX-related factors {3.1.1} | ||||||||
| TF subfamily | EVX (Even-skipped homolog) {3.1.1.10} | ||||||||
| HGNC | HGNC:3507 | ||||||||
| EntrezGene | GeneID:344191 (SSTAR profile) | ||||||||
| UniProt ID | EVX2_HUMAN | ||||||||
| UniProt AC | Q03828 (TFClass) | ||||||||
| Comment | Retracted motif! | ||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | EVX2 expression | ||||||||
| ReMap ChIP-seq dataset list | EVX2 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 43.181 | 21.074 | 31.196 | 29.735 | 38.897 | 23.059 | 5.108 | 51.785 | 26.276 | 14.371 | 8.928 | 28.949 | 37.093 | 29.482 | 35.007 | 69.289 |
| 02 | 42.055 | 18.634 | 40.702 | 44.057 | 34.841 | 11.346 | 3.744 | 38.054 | 22.808 | 15.131 | 16.993 | 25.307 | 27.273 | 21.595 | 45.793 | 85.096 |
| 03 | 39.832 | 24.566 | 39.583 | 22.997 | 25.397 | 14.036 | 7.619 | 19.653 | 26.05 | 25.883 | 27.928 | 27.371 | 34.989 | 47.874 | 67.921 | 41.728 |
| 04 | 56.722 | 25.541 | 20.052 | 23.953 | 29.483 | 29.363 | 3.075 | 50.437 | 46.783 | 35.168 | 26.11 | 34.99 | 24.309 | 14.838 | 20.304 | 52.299 |
| 05 | 38.213 | 43.796 | 37.07 | 38.219 | 40.602 | 31.538 | 5.163 | 27.608 | 23.711 | 18.023 | 14.355 | 13.451 | 28.302 | 38.385 | 48.133 | 46.86 |
| 06 | 50.305 | 26.77 | 29.908 | 23.846 | 53.623 | 25.32 | 3.271 | 49.527 | 35.899 | 22.042 | 20.365 | 26.414 | 21.171 | 21.215 | 22.024 | 61.729 |
| 07 | 23.261 | 25.196 | 43.18 | 69.361 | 31.99 | 16.188 | 6.378 | 40.791 | 11.039 | 15.055 | 29.046 | 20.429 | 22.705 | 31.036 | 71.715 | 36.059 |
| 08 | 8.885 | 17.048 | 19.559 | 43.503 | 27.029 | 19.142 | 6.042 | 35.26 | 35.823 | 25.32 | 25.221 | 63.955 | 21.74 | 57.443 | 37.789 | 49.667 |
| 09 | 0.898 | 0.0 | 1.864 | 90.715 | 1.062 | 0.997 | 2.836 | 114.059 | 1.03 | 0.906 | 1.962 | 84.714 | 0.0 | 0.927 | 1.099 | 190.359 |
| 10 | 0.0 | 0.0 | 0.0 | 2.989 | 0.0 | 0.0 | 0.0 | 2.83 | 0.0 | 0.0 | 1.018 | 6.744 | 5.073 | 4.046 | 1.2 | 469.527 |
| 11 | 0.0 | 0.0 | 0.0 | 5.073 | 0.0 | 1.046 | 0.0 | 3.0 | 1.018 | 0.0 | 1.2 | 0.0 | 21.541 | 24.28 | 8.76 | 427.51 |
| 12 | 20.207 | 0.0 | 0.0 | 2.352 | 16.462 | 5.026 | 0.0 | 3.838 | 7.109 | 1.923 | 0.0 | 0.927 | 397.766 | 8.976 | 16.364 | 12.478 |
| 13 | 3.048 | 71.388 | 15.777 | 351.332 | 0.0 | 1.926 | 0.0 | 14.0 | 0.984 | 1.025 | 0.909 | 13.445 | 0.0 | 2.836 | 0.0 | 16.759 |
| 14 | 1.861 | 0.0 | 1.187 | 0.984 | 50.268 | 0.971 | 3.334 | 22.603 | 0.0 | 1.038 | 7.036 | 8.613 | 28.201 | 3.678 | 169.312 | 194.344 |
| 15 | 18.514 | 5.887 | 52.872 | 3.057 | 2.776 | 1.873 | 0.0 | 1.038 | 66.954 | 7.405 | 102.476 | 4.033 | 51.389 | 18.496 | 143.598 | 13.061 |
| 16 | 4.851 | 101.833 | 17.392 | 15.557 | 1.956 | 28.609 | 1.05 | 2.045 | 24.015 | 197.422 | 23.747 | 53.762 | 0.944 | 15.439 | 1.071 | 3.736 |
| 17 | 2.016 | 12.87 | 10.961 | 5.918 | 69.647 | 125.571 | 5.083 | 143.002 | 11.873 | 21.917 | 4.418 | 5.051 | 11.73 | 18.847 | 20.926 | 23.597 |
| 18 | 9.46 | 45.22 | 25.636 | 14.951 | 23.155 | 70.487 | 14.552 | 71.012 | 3.01 | 17.728 | 10.81 | 9.839 | 16.811 | 34.556 | 64.114 | 62.088 |
| 19 | 12.802 | 12.496 | 7.044 | 20.095 | 46.558 | 49.592 | 7.025 | 64.815 | 13.764 | 26.418 | 34.383 | 40.546 | 16.789 | 34.452 | 32.011 | 74.638 |
| 20 | 20.166 | 23.593 | 30.036 | 16.119 | 39.465 | 45.224 | 2.352 | 35.917 | 13.067 | 19.328 | 22.933 | 25.135 | 14.485 | 47.808 | 63.099 | 74.702 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.333 | -0.375 | 0.011 | -0.036 | 0.23 | -0.287 | -1.737 | 0.513 | -0.158 | -0.749 | -1.21 | -0.062 | 0.183 | -0.044 | 0.125 | 0.803 |
| 02 | 0.307 | -0.496 | 0.274 | 0.353 | 0.121 | -0.979 | -2.023 | 0.208 | -0.297 | -0.699 | -0.586 | -0.195 | -0.121 | -0.351 | 0.391 | 1.007 |
| 03 | 0.253 | -0.224 | 0.247 | -0.289 | -0.191 | -0.772 | -1.361 | -0.443 | -0.166 | -0.173 | -0.098 | -0.118 | 0.125 | 0.435 | 0.783 | 0.299 |
| 04 | 0.604 | -0.186 | -0.424 | -0.249 | -0.044 | -0.048 | -2.199 | 0.487 | 0.413 | 0.13 | -0.164 | 0.125 | -0.235 | -0.718 | -0.412 | 0.523 |
| 05 | 0.212 | 0.347 | 0.182 | 0.212 | 0.272 | 0.022 | -1.727 | -0.109 | -0.259 | -0.528 | -0.751 | -0.814 | -0.085 | 0.216 | 0.441 | 0.414 |
| 06 | 0.484 | -0.14 | -0.03 | -0.254 | 0.548 | -0.195 | -2.144 | 0.469 | 0.15 | -0.331 | -0.409 | -0.153 | -0.371 | -0.368 | -0.332 | 0.688 |
| 07 | -0.278 | -0.199 | 0.333 | 0.804 | 0.036 | -0.633 | -1.529 | 0.277 | -1.005 | -0.704 | -0.059 | -0.406 | -0.302 | 0.006 | 0.837 | 0.155 |
| 08 | -1.214 | -0.583 | -0.448 | 0.34 | -0.13 | -0.469 | -1.58 | 0.132 | 0.148 | -0.194 | -0.198 | 0.723 | -0.344 | 0.616 | 0.201 | 0.472 |
| 09 | -3.19 | -4.389 | -2.629 | 1.071 | -3.07 | -3.116 | -2.271 | 1.299 | -3.093 | -3.184 | -2.587 | 1.003 | -4.389 | -3.168 | -3.045 | 1.81 |
| 10 | -4.389 | -4.389 | -4.389 | -2.224 | -4.389 | -4.389 | -4.389 | -2.273 | -4.389 | -4.389 | -3.101 | -1.477 | -1.744 | -1.952 | -2.979 | 2.711 |
| 11 | -4.389 | -4.389 | -4.389 | -1.744 | -4.389 | -3.081 | -4.389 | -2.221 | -3.101 | -4.389 | -2.979 | -4.389 | -0.354 | -0.236 | -1.228 | 2.618 |
| 12 | -0.416 | -4.389 | -4.389 | -2.434 | -0.617 | -1.752 | -4.389 | -2.0 | -1.427 | -2.604 | -4.389 | -3.168 | 2.546 | -1.204 | -0.623 | -0.887 |
| 13 | -2.207 | 0.832 | -0.658 | 2.422 | -4.389 | -2.603 | -4.389 | -0.775 | -3.125 | -3.096 | -3.181 | -0.814 | -4.389 | -2.271 | -4.389 | -0.599 |
| 14 | -2.631 | -4.389 | -2.987 | -3.125 | 0.484 | -3.135 | -2.127 | -0.306 | -4.389 | -3.087 | -1.437 | -1.244 | -0.088 | -2.039 | 1.693 | 1.83 |
| 15 | -0.502 | -1.605 | 0.534 | -2.204 | -2.29 | -2.626 | -4.389 | -3.087 | 0.769 | -1.388 | 1.192 | -1.955 | 0.506 | -0.503 | 1.528 | -0.842 |
| 16 | -1.785 | 1.186 | -0.563 | -0.672 | -2.59 | -0.074 | -3.078 | -2.552 | -0.247 | 1.846 | -0.258 | 0.55 | -3.155 | -0.68 | -3.064 | -2.025 |
| 17 | -2.564 | -0.857 | -1.012 | -1.6 | 0.808 | 1.395 | -1.742 | 1.524 | -0.935 | -0.336 | -1.871 | -1.748 | -0.947 | -0.485 | -0.382 | -0.264 |
| 18 | -1.154 | 0.379 | -0.182 | -0.711 | -0.282 | 0.82 | -0.737 | 0.827 | -2.218 | -0.545 | -1.026 | -1.116 | -0.596 | 0.112 | 0.725 | 0.693 |
| 19 | -0.862 | -0.885 | -1.436 | -0.422 | 0.408 | 0.47 | -1.438 | 0.736 | -0.791 | -0.153 | 0.107 | 0.271 | -0.598 | 0.109 | 0.037 | 0.877 |
| 20 | -0.418 | -0.264 | -0.026 | -0.638 | 0.244 | 0.379 | -2.433 | 0.151 | -0.842 | -0.46 | -0.292 | -0.202 | -0.742 | 0.434 | 0.71 | 0.877 |