Model info
| Transcription factor | Onecut1 | ||||||||
| Model | HNF6_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 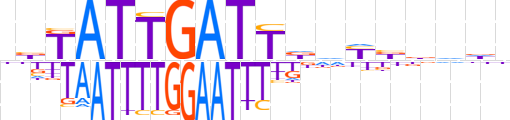 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dbTATTGATTddYbdddn | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.977 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 24 | ||||||||
| Aligned words | 306 | ||||||||
| TF family | HD-CUT factors {3.1.9} | ||||||||
| TF subfamily | ONECUT {3.1.9.1} | ||||||||
| MGI | MGI:1196423 | ||||||||
| EntrezGene | GeneID:15379 (SSTAR profile) | ||||||||
| UniProt ID | HNF6_MOUSE | ||||||||
| UniProt AC | O08755 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Onecut1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 11.0 | 10.0 | 31.0 | 22.0 | 4.0 | 12.0 | 2.0 | 19.0 | 6.0 | 11.0 | 10.0 | 23.0 | 9.0 | 11.0 | 22.0 | 82.0 |
| 02 | 0.0 | 1.0 | 1.0 | 28.0 | 0.0 | 1.0 | 2.0 | 41.0 | 0.0 | 7.0 | 4.0 | 54.0 | 4.0 | 9.0 | 28.0 | 105.0 |
| 03 | 4.0 | 0.0 | 0.0 | 0.0 | 18.0 | 0.0 | 0.0 | 0.0 | 35.0 | 0.0 | 0.0 | 0.0 | 228.0 | 0.0 | 0.0 | 0.0 |
| 04 | 0.0 | 0.0 | 0.0 | 285.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 05 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 28.0 | 0.0 | 257.0 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 28.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 257.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 285.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 285.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 57.0 | 3.0 | 225.0 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 7.0 | 11.0 | 13.0 | 26.0 | 0.0 | 0.0 | 1.0 | 2.0 | 28.0 | 9.0 | 83.0 | 105.0 |
| 11 | 11.0 | 4.0 | 9.0 | 11.0 | 13.0 | 6.0 | 0.0 | 1.0 | 71.0 | 8.0 | 10.0 | 8.0 | 19.0 | 25.0 | 36.0 | 53.0 |
| 12 | 10.0 | 19.0 | 5.0 | 80.0 | 7.0 | 12.0 | 0.0 | 24.0 | 10.0 | 3.0 | 7.0 | 35.0 | 10.0 | 15.0 | 6.0 | 42.0 |
| 13 | 4.0 | 8.0 | 11.0 | 14.0 | 13.0 | 12.0 | 1.0 | 23.0 | 2.0 | 4.0 | 5.0 | 7.0 | 13.0 | 26.0 | 33.0 | 109.0 |
| 14 | 7.0 | 2.0 | 14.0 | 9.0 | 26.0 | 4.0 | 3.0 | 17.0 | 11.0 | 12.0 | 19.0 | 8.0 | 13.0 | 11.0 | 63.0 | 66.0 |
| 15 | 25.0 | 13.0 | 10.0 | 9.0 | 12.0 | 6.0 | 1.0 | 10.0 | 57.0 | 13.0 | 15.0 | 14.0 | 19.0 | 15.0 | 32.0 | 34.0 |
| 16 | 27.0 | 10.0 | 18.0 | 58.0 | 17.0 | 5.0 | 1.0 | 24.0 | 14.0 | 8.0 | 17.0 | 19.0 | 5.0 | 14.0 | 10.0 | 38.0 |
| 17 | 21.0 | 6.0 | 10.0 | 26.0 | 10.0 | 11.0 | 0.0 | 16.0 | 4.0 | 12.0 | 14.0 | 16.0 | 13.0 | 20.0 | 24.0 | 82.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.47 | -0.562 | 0.546 | 0.207 | -1.429 | -0.386 | -2.044 | 0.063 | -1.051 | -0.47 | -0.562 | 0.251 | -0.664 | -0.47 | 0.207 | 1.511 |
| 02 | -3.94 | -2.597 | -2.597 | 0.445 | -3.94 | -2.597 | -2.044 | 0.823 | -3.94 | -0.904 | -1.429 | 1.096 | -1.429 | -0.664 | 0.445 | 1.758 |
| 03 | -1.429 | -3.94 | -3.94 | -3.94 | 0.01 | -3.94 | -3.94 | -3.94 | 0.666 | -3.94 | -3.94 | -3.94 | 2.531 | -3.94 | -3.94 | -3.94 |
| 04 | -3.94 | -3.94 | -3.94 | 2.754 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 |
| 05 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | 0.445 | -3.94 | 2.651 |
| 06 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | 0.445 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | 2.651 | -3.94 |
| 07 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | 2.754 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 |
| 08 | -3.94 | -3.94 | -3.94 | 2.754 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 |
| 09 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | -3.94 | 1.15 | -1.69 | 2.518 |
| 10 | -3.94 | -3.94 | -3.94 | -3.94 | -0.904 | -0.47 | -0.308 | 0.372 | -3.94 | -3.94 | -2.597 | -2.044 | 0.445 | -0.664 | 1.524 | 1.758 |
| 11 | -0.47 | -1.429 | -0.664 | -0.47 | -0.308 | -1.051 | -3.94 | -2.597 | 1.368 | -0.777 | -0.562 | -0.777 | 0.063 | 0.333 | 0.694 | 1.077 |
| 12 | -0.562 | 0.063 | -1.222 | 1.487 | -0.904 | -0.386 | -3.94 | 0.293 | -0.562 | -1.69 | -0.904 | 0.666 | -0.562 | -0.168 | -1.051 | 0.847 |
| 13 | -1.429 | -0.777 | -0.47 | -0.236 | -0.308 | -0.386 | -2.597 | 0.251 | -2.044 | -1.429 | -1.222 | -0.904 | -0.308 | 0.372 | 0.608 | 1.795 |
| 14 | -0.904 | -2.044 | -0.236 | -0.664 | 0.372 | -1.429 | -1.69 | -0.046 | -0.47 | -0.386 | 0.063 | -0.777 | -0.308 | -0.47 | 1.249 | 1.295 |
| 15 | 0.333 | -0.308 | -0.562 | -0.664 | -0.386 | -1.051 | -2.597 | -0.562 | 1.15 | -0.308 | -0.168 | -0.236 | 0.063 | -0.168 | 0.577 | 0.637 |
| 16 | 0.409 | -0.562 | 0.01 | 1.167 | -0.046 | -1.222 | -2.597 | 0.293 | -0.236 | -0.777 | -0.046 | 0.063 | -1.222 | -0.236 | -0.562 | 0.747 |
| 17 | 0.162 | -1.051 | -0.562 | 0.372 | -0.562 | -0.47 | -3.94 | -0.105 | -1.429 | -0.386 | -0.236 | -0.105 | -0.308 | 0.114 | 0.293 | 1.511 |