Model info
| Transcription factor | Nfic | ||||||||
| Model | NFIC_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 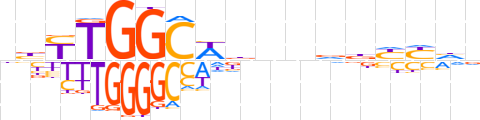 | ||||||||
| LOGO (reverse complement) | 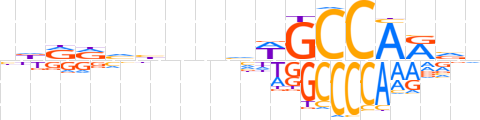 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 17 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbYTGGCWbnndbYYhd | ||||||||
| Best auROC (human) | 0.976 | ||||||||
| Best auROC (mouse) | 0.879 | ||||||||
| Peak sets in benchmark (human) | 18 | ||||||||
| Peak sets in benchmark (mouse) | 1 | ||||||||
| Aligned words | 295 | ||||||||
| TF family | Nuclear factor 1 {7.1.2} | ||||||||
| TF subfamily | NF-1C {7.1.2.0.3} | ||||||||
| MGI | MGI:109591 | ||||||||
| EntrezGene | GeneID:18029 (SSTAR profile) | ||||||||
| UniProt ID | NFIC_MOUSE | ||||||||
| UniProt AC | P70255 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Nfic expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 4.0 | 26.0 | 19.0 | 42.0 | 2.0 | 16.0 | 5.0 | 23.0 | 5.0 | 24.0 | 4.0 | 13.0 | 7.0 | 49.0 | 22.0 | 34.0 |
| 02 | 0.0 | 8.0 | 1.0 | 9.0 | 0.0 | 32.0 | 0.0 | 83.0 | 0.0 | 28.0 | 1.0 | 21.0 | 2.0 | 28.0 | 10.0 | 72.0 |
| 03 | 0.0 | 0.0 | 0.0 | 2.0 | 5.0 | 0.0 | 3.0 | 88.0 | 1.0 | 0.0 | 1.0 | 10.0 | 2.0 | 0.0 | 25.0 | 158.0 |
| 04 | 0.0 | 0.0 | 8.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 29.0 | 0.0 | 0.0 | 0.0 | 257.0 | 1.0 |
| 05 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 281.0 | 13.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 40.0 | 242.0 | 0.0 | 0.0 | 1.0 | 12.0 | 0.0 | 0.0 |
| 07 | 32.0 | 1.0 | 1.0 | 7.0 | 141.0 | 17.0 | 7.0 | 89.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 25.0 | 31.0 | 77.0 | 40.0 | 6.0 | 4.0 | 1.0 | 7.0 | 0.0 | 4.0 | 2.0 | 2.0 | 4.0 | 22.0 | 33.0 | 37.0 |
| 09 | 12.0 | 6.0 | 12.0 | 5.0 | 24.0 | 15.0 | 1.0 | 21.0 | 34.0 | 28.0 | 28.0 | 23.0 | 15.0 | 16.0 | 29.0 | 26.0 |
| 10 | 25.0 | 12.0 | 35.0 | 13.0 | 24.0 | 15.0 | 12.0 | 14.0 | 26.0 | 16.0 | 17.0 | 11.0 | 13.0 | 10.0 | 33.0 | 19.0 |
| 11 | 29.0 | 8.0 | 42.0 | 9.0 | 22.0 | 9.0 | 5.0 | 17.0 | 40.0 | 9.0 | 37.0 | 11.0 | 17.0 | 3.0 | 17.0 | 20.0 |
| 12 | 15.0 | 13.0 | 53.0 | 27.0 | 3.0 | 9.0 | 3.0 | 14.0 | 14.0 | 17.0 | 38.0 | 32.0 | 2.0 | 7.0 | 38.0 | 10.0 |
| 13 | 16.0 | 11.0 | 3.0 | 4.0 | 9.0 | 18.0 | 2.0 | 17.0 | 7.0 | 94.0 | 11.0 | 20.0 | 6.0 | 48.0 | 4.0 | 25.0 |
| 14 | 7.0 | 12.0 | 7.0 | 12.0 | 17.0 | 140.0 | 1.0 | 13.0 | 3.0 | 6.0 | 2.0 | 9.0 | 8.0 | 23.0 | 7.0 | 28.0 |
| 15 | 8.0 | 6.0 | 12.0 | 9.0 | 124.0 | 37.0 | 3.0 | 17.0 | 3.0 | 6.0 | 5.0 | 3.0 | 18.0 | 7.0 | 19.0 | 18.0 |
| 16 | 46.0 | 16.0 | 69.0 | 22.0 | 31.0 | 8.0 | 3.0 | 14.0 | 8.0 | 12.0 | 14.0 | 5.0 | 11.0 | 5.0 | 14.0 | 17.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.462 | 0.338 | 0.029 | 0.813 | -2.077 | -0.139 | -1.255 | 0.217 | -1.255 | 0.259 | -1.462 | -0.342 | -0.938 | 0.966 | 0.174 | 0.603 |
| 02 | -3.968 | -0.811 | -2.629 | -0.698 | -3.968 | 0.543 | -3.968 | 1.49 | -3.968 | 0.411 | -2.629 | 0.128 | -2.077 | 0.411 | -0.596 | 1.348 |
| 03 | -3.968 | -3.968 | -3.968 | -2.077 | -1.255 | -3.968 | -1.723 | 1.548 | -2.629 | -3.968 | -2.629 | -0.596 | -2.077 | -3.968 | 0.3 | 2.131 |
| 04 | -3.968 | -3.968 | -0.811 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | 0.446 | -3.968 | -3.968 | -3.968 | 2.617 | -2.629 |
| 05 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | 2.706 | -0.342 | -3.968 | -3.968 | -2.629 | -3.968 |
| 06 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | 0.764 | 2.557 | -3.968 | -3.968 | -2.629 | -0.419 | -3.968 | -3.968 |
| 07 | 0.543 | -2.629 | -2.629 | -0.938 | 2.018 | -0.08 | -0.938 | 1.559 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 | -3.968 |
| 08 | 0.3 | 0.512 | 1.415 | 0.764 | -1.084 | -1.462 | -2.629 | -0.938 | -3.968 | -1.462 | -2.077 | -2.077 | -1.462 | 0.174 | 0.574 | 0.687 |
| 09 | -0.419 | -1.084 | -0.419 | -1.255 | 0.259 | -0.202 | -2.629 | 0.128 | 0.603 | 0.411 | 0.411 | 0.217 | -0.202 | -0.139 | 0.446 | 0.338 |
| 10 | 0.3 | -0.419 | 0.632 | -0.342 | 0.259 | -0.202 | -0.419 | -0.269 | 0.338 | -0.139 | -0.08 | -0.504 | -0.342 | -0.596 | 0.574 | 0.029 |
| 11 | 0.446 | -0.811 | 0.813 | -0.698 | 0.174 | -0.698 | -1.255 | -0.08 | 0.764 | -0.698 | 0.687 | -0.504 | -0.08 | -1.723 | -0.08 | 0.08 |
| 12 | -0.202 | -0.342 | 1.043 | 0.375 | -1.723 | -0.698 | -1.723 | -0.269 | -0.269 | -0.08 | 0.713 | 0.543 | -2.077 | -0.938 | 0.713 | -0.596 |
| 13 | -0.139 | -0.504 | -1.723 | -1.462 | -0.698 | -0.024 | -2.077 | -0.08 | -0.938 | 1.614 | -0.504 | 0.08 | -1.084 | 0.945 | -1.462 | 0.3 |
| 14 | -0.938 | -0.419 | -0.938 | -0.419 | -0.08 | 2.011 | -2.629 | -0.342 | -1.723 | -1.084 | -2.077 | -0.698 | -0.811 | 0.217 | -0.938 | 0.411 |
| 15 | -0.811 | -1.084 | -0.419 | -0.698 | 1.89 | 0.687 | -1.723 | -0.08 | -1.723 | -1.084 | -1.255 | -1.723 | -0.024 | -0.938 | 0.029 | -0.024 |
| 16 | 0.903 | -0.139 | 1.306 | 0.174 | 0.512 | -0.811 | -1.723 | -0.269 | -0.811 | -0.419 | -0.269 | -1.255 | -0.504 | -1.255 | -0.269 | -0.08 |