Model info
| Transcription factor | SIX1 (GeneCards) | ||||||||
| Model | SIX1_HUMAN.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 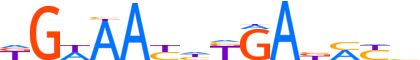 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 14 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | KGWAAYhKGAbMYb | ||||||||
| Best auROC (human) | 0.961 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 6 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 500 | ||||||||
| TF family | HD-SINE factors {3.1.6} | ||||||||
| TF subfamily | SIX1-like factors {3.1.6.1} | ||||||||
| HGNC | HGNC:10887 | ||||||||
| EntrezGene | GeneID:6495 (SSTAR profile) | ||||||||
| UniProt ID | SIX1_HUMAN | ||||||||
| UniProt AC | Q15475 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | SIX1 expression | ||||||||
| ReMap ChIP-seq dataset list | SIX1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 98.0 | 9.0 | 103.0 | 290.0 |
| 02 | 8.0 | 2.0 | 489.0 | 1.0 |
| 03 | 236.0 | 22.0 | 24.0 | 218.0 |
| 04 | 451.0 | 24.0 | 1.0 | 24.0 |
| 05 | 496.0 | 2.0 | 1.0 | 1.0 |
| 06 | 21.0 | 247.0 | 24.0 | 208.0 |
| 07 | 109.0 | 196.0 | 23.0 | 172.0 |
| 08 | 44.0 | 51.0 | 55.0 | 350.0 |
| 09 | 52.0 | 7.0 | 423.0 | 18.0 |
| 10 | 498.0 | 0.0 | 1.0 | 1.0 |
| 11 | 38.0 | 74.0 | 153.0 | 235.0 |
| 12 | 255.0 | 211.0 | 12.0 | 22.0 |
| 13 | 42.0 | 294.0 | 20.0 | 144.0 |
| 14 | 69.0 | 198.0 | 95.0 | 138.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.24 | -2.484 | -0.191 | 0.835 |
| 02 | -2.584 | -3.573 | 1.355 | -3.903 |
| 03 | 0.63 | -1.681 | -1.6 | 0.551 |
| 04 | 1.274 | -1.6 | -3.903 | -1.6 |
| 05 | 1.369 | -3.573 | -3.903 | -3.903 |
| 06 | -1.725 | 0.675 | -1.6 | 0.504 |
| 07 | -0.135 | 0.445 | -1.64 | 0.316 |
| 08 | -1.022 | -0.879 | -0.805 | 1.022 |
| 09 | -0.86 | -2.694 | 1.21 | -1.868 |
| 10 | 1.373 | -4.4 | -3.903 | -3.903 |
| 11 | -1.163 | -0.516 | 0.2 | 0.626 |
| 12 | 0.707 | 0.519 | -2.234 | -1.681 |
| 13 | -1.067 | 0.848 | -1.77 | 0.14 |
| 14 | -0.584 | 0.455 | -0.271 | 0.098 |