Model info
| Transcription factor | Tcf3 | ||||||||
| Model | TFE2_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 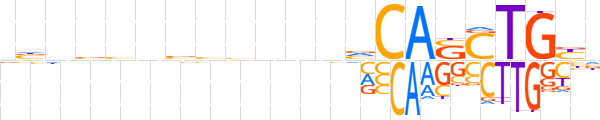 | ||||||||
| LOGO (reverse complement) | 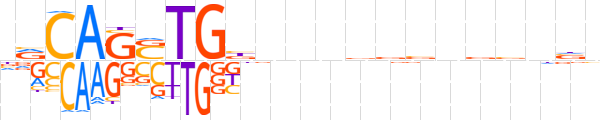 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 21 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbnbbnbbhbnnvCASCTGYn | ||||||||
| Best auROC (human) | 0.955 | ||||||||
| Best auROC (mouse) | 0.987 | ||||||||
| Peak sets in benchmark (human) | 9 | ||||||||
| Peak sets in benchmark (mouse) | 60 | ||||||||
| Aligned words | 467 | ||||||||
| TF family | E2A-related factors {1.2.1} | ||||||||
| TF subfamily | E2A (TCF-3, ITF-1) {1.2.1.0.1} | ||||||||
| MGI | MGI:98510 | ||||||||
| EntrezGene | GeneID:21423 (SSTAR profile) | ||||||||
| UniProt ID | TFE2_MOUSE | ||||||||
| UniProt AC | P15806 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Tcf3 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 13.0 | 36.0 | 19.0 | 17.0 | 32.0 | 71.0 | 11.0 | 31.0 | 13.0 | 85.0 | 26.0 | 16.0 | 8.0 | 45.0 | 23.0 | 4.0 |
| 02 | 13.0 | 20.0 | 19.0 | 14.0 | 88.0 | 58.0 | 11.0 | 80.0 | 16.0 | 27.0 | 12.0 | 24.0 | 6.0 | 29.0 | 23.0 | 10.0 |
| 03 | 6.0 | 53.0 | 56.0 | 8.0 | 40.0 | 57.0 | 6.0 | 31.0 | 8.0 | 20.0 | 21.0 | 16.0 | 10.0 | 30.0 | 72.0 | 16.0 |
| 04 | 8.0 | 19.0 | 24.0 | 13.0 | 43.0 | 69.0 | 14.0 | 34.0 | 12.0 | 77.0 | 34.0 | 32.0 | 6.0 | 29.0 | 26.0 | 10.0 |
| 05 | 17.0 | 12.0 | 19.0 | 21.0 | 56.0 | 46.0 | 16.0 | 76.0 | 19.0 | 30.0 | 22.0 | 27.0 | 11.0 | 25.0 | 23.0 | 30.0 |
| 06 | 10.0 | 36.0 | 46.0 | 11.0 | 25.0 | 45.0 | 15.0 | 28.0 | 6.0 | 41.0 | 12.0 | 21.0 | 5.0 | 50.0 | 71.0 | 28.0 |
| 07 | 13.0 | 20.0 | 11.0 | 2.0 | 39.0 | 94.0 | 9.0 | 30.0 | 18.0 | 60.0 | 41.0 | 25.0 | 4.0 | 32.0 | 26.0 | 26.0 |
| 08 | 20.0 | 29.0 | 19.0 | 6.0 | 68.0 | 69.0 | 13.0 | 56.0 | 13.0 | 34.0 | 24.0 | 16.0 | 11.0 | 47.0 | 16.0 | 9.0 |
| 09 | 12.0 | 39.0 | 53.0 | 8.0 | 43.0 | 63.0 | 15.0 | 58.0 | 7.0 | 26.0 | 29.0 | 10.0 | 6.0 | 34.0 | 42.0 | 5.0 |
| 10 | 14.0 | 19.0 | 23.0 | 12.0 | 49.0 | 63.0 | 5.0 | 45.0 | 30.0 | 52.0 | 24.0 | 33.0 | 5.0 | 31.0 | 29.0 | 16.0 |
| 11 | 10.0 | 43.0 | 33.0 | 12.0 | 46.0 | 46.0 | 11.0 | 62.0 | 26.0 | 34.0 | 15.0 | 6.0 | 14.0 | 46.0 | 40.0 | 6.0 |
| 12 | 17.0 | 34.0 | 43.0 | 2.0 | 88.0 | 52.0 | 18.0 | 11.0 | 31.0 | 48.0 | 20.0 | 0.0 | 14.0 | 32.0 | 39.0 | 1.0 |
| 13 | 0.0 | 147.0 | 2.0 | 1.0 | 0.0 | 164.0 | 0.0 | 2.0 | 0.0 | 119.0 | 1.0 | 0.0 | 0.0 | 13.0 | 1.0 | 0.0 |
| 14 | 0.0 | 0.0 | 0.0 | 0.0 | 433.0 | 3.0 | 3.0 | 4.0 | 3.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 |
| 15 | 0.0 | 127.0 | 275.0 | 37.0 | 1.0 | 2.0 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 3.0 | 0.0 | 1.0 |
| 16 | 0.0 | 1.0 | 0.0 | 0.0 | 9.0 | 112.0 | 9.0 | 3.0 | 16.0 | 229.0 | 32.0 | 0.0 | 11.0 | 23.0 | 5.0 | 0.0 |
| 17 | 0.0 | 0.0 | 1.0 | 35.0 | 1.0 | 2.0 | 3.0 | 359.0 | 0.0 | 0.0 | 2.0 | 44.0 | 0.0 | 0.0 | 1.0 | 2.0 |
| 18 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 2.0 | 5.0 | 0.0 | 2.0 | 4.0 | 417.0 | 17.0 |
| 19 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 4.0 | 0.0 | 1.0 | 23.0 | 232.0 | 58.0 | 112.0 | 0.0 | 13.0 | 1.0 | 3.0 |
| 20 | 3.0 | 4.0 | 17.0 | 0.0 | 93.0 | 73.0 | 21.0 | 63.0 | 0.0 | 26.0 | 17.0 | 16.0 | 7.0 | 32.0 | 69.0 | 9.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.756 | 0.244 | -0.386 | -0.495 | 0.127 | 0.918 | -0.918 | 0.096 | -0.756 | 1.097 | -0.077 | -0.554 | -1.224 | 0.465 | -0.198 | -1.873 |
| 02 | -0.756 | -0.335 | -0.386 | -0.684 | 1.132 | 0.717 | -0.918 | 1.037 | -0.554 | -0.04 | -0.834 | -0.156 | -1.497 | 0.03 | -0.198 | -1.01 |
| 03 | -1.497 | 0.627 | 0.682 | -1.224 | 0.348 | 0.7 | -1.497 | 0.096 | -1.224 | -0.335 | -0.288 | -0.554 | -1.01 | 0.064 | 0.932 | -0.554 |
| 04 | -1.224 | -0.386 | -0.156 | -0.756 | 0.42 | 0.889 | -0.684 | 0.187 | -0.834 | 0.999 | 0.187 | 0.127 | -1.497 | 0.03 | -0.077 | -1.01 |
| 05 | -0.495 | -0.834 | -0.386 | -0.288 | 0.682 | 0.487 | -0.554 | 0.986 | -0.386 | 0.064 | -0.242 | -0.04 | -0.918 | -0.116 | -0.198 | 0.064 |
| 06 | -1.01 | 0.244 | 0.487 | -0.918 | -0.116 | 0.465 | -0.617 | -0.004 | -1.497 | 0.373 | -0.834 | -0.288 | -1.667 | 0.569 | 0.918 | -0.004 |
| 07 | -0.756 | -0.335 | -0.918 | -2.482 | 0.323 | 1.197 | -1.111 | 0.064 | -0.439 | 0.751 | 0.373 | -0.116 | -1.873 | 0.127 | -0.077 | -0.077 |
| 08 | -0.335 | 0.03 | -0.386 | -1.497 | 0.875 | 0.889 | -0.756 | 0.682 | -0.756 | 0.187 | -0.156 | -0.554 | -0.918 | 0.508 | -0.554 | -1.111 |
| 09 | -0.834 | 0.323 | 0.627 | -1.224 | 0.42 | 0.799 | -0.617 | 0.717 | -1.351 | -0.077 | 0.03 | -1.01 | -1.497 | 0.187 | 0.397 | -1.667 |
| 10 | -0.684 | -0.386 | -0.198 | -0.834 | 0.549 | 0.799 | -1.667 | 0.465 | 0.064 | 0.608 | -0.156 | 0.158 | -1.667 | 0.096 | 0.03 | -0.554 |
| 11 | -1.01 | 0.42 | 0.158 | -0.834 | 0.487 | 0.487 | -0.918 | 0.783 | -0.077 | 0.187 | -0.617 | -1.497 | -0.684 | 0.487 | 0.348 | -1.497 |
| 12 | -0.495 | 0.187 | 0.42 | -2.482 | 1.132 | 0.608 | -0.439 | -0.918 | 0.096 | 0.529 | -0.335 | -4.313 | -0.684 | 0.127 | 0.323 | -3.027 |
| 13 | -4.313 | 1.643 | -2.482 | -3.027 | -4.313 | 1.752 | -4.313 | -2.482 | -4.313 | 1.432 | -3.027 | -4.313 | -4.313 | -0.756 | -3.027 | -4.313 |
| 14 | -4.313 | -4.313 | -4.313 | -4.313 | 2.721 | -2.132 | -2.132 | -1.873 | -2.132 | -3.027 | -4.313 | -4.313 | -2.132 | -4.313 | -4.313 | -4.313 |
| 15 | -4.313 | 1.497 | 2.268 | 0.271 | -3.027 | -2.482 | -3.027 | -4.313 | -4.313 | -3.027 | -3.027 | -3.027 | -4.313 | -2.132 | -4.313 | -3.027 |
| 16 | -4.313 | -3.027 | -4.313 | -4.313 | -1.111 | 1.372 | -1.111 | -2.132 | -0.554 | 2.085 | 0.127 | -4.313 | -0.918 | -0.198 | -1.667 | -4.313 |
| 17 | -4.313 | -4.313 | -3.027 | 0.216 | -3.027 | -2.482 | -2.132 | 2.534 | -4.313 | -4.313 | -2.482 | 0.443 | -4.313 | -4.313 | -3.027 | -2.482 |
| 18 | -4.313 | -4.313 | -3.027 | -4.313 | -4.313 | -4.313 | -2.482 | -4.313 | -4.313 | -2.482 | -1.667 | -4.313 | -2.482 | -1.873 | 2.684 | -0.495 |
| 19 | -4.313 | -3.027 | -4.313 | -3.027 | -3.027 | -1.873 | -4.313 | -3.027 | -0.198 | 2.098 | 0.717 | 1.372 | -4.313 | -0.756 | -3.027 | -2.132 |
| 20 | -2.132 | -1.873 | -0.495 | -4.313 | 1.187 | 0.946 | -0.288 | 0.799 | -4.313 | -0.077 | -0.495 | -0.554 | -1.351 | 0.127 | 0.889 | -1.111 |