Model info
| Transcription factor | ZNF263 (GeneCards) | ||||||||
| Model | ZN263_HUMAN.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 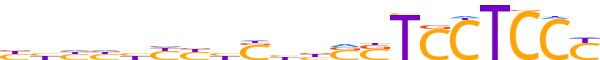 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 20 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | RGGAGGASSvvSvRRvvvnv | ||||||||
| Best auROC (human) | 0.968 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 9 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 501 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:13056 | ||||||||
| EntrezGene | GeneID:10127 (SSTAR profile) | ||||||||
| UniProt ID | ZN263_HUMAN | ||||||||
| UniProt AC | O14978 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF263 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF263 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 101.0 | 17.0 | 355.0 | 26.0 |
| 02 | 12.0 | 10.0 | 459.0 | 18.0 |
| 03 | 9.0 | 11.0 | 478.0 | 1.0 |
| 04 | 491.0 | 7.0 | 0.0 | 1.0 |
| 05 | 24.0 | 1.0 | 454.0 | 20.0 |
| 06 | 13.0 | 55.0 | 423.0 | 8.0 |
| 07 | 468.0 | 4.0 | 23.0 | 4.0 |
| 08 | 52.0 | 105.0 | 317.0 | 25.0 |
| 09 | 22.0 | 83.0 | 314.0 | 80.0 |
| 10 | 171.0 | 80.0 | 212.0 | 36.0 |
| 11 | 208.0 | 75.0 | 170.0 | 46.0 |
| 12 | 55.0 | 57.0 | 358.0 | 29.0 |
| 13 | 206.0 | 55.0 | 195.0 | 43.0 |
| 14 | 116.0 | 54.0 | 295.0 | 34.0 |
| 15 | 94.0 | 46.0 | 310.0 | 49.0 |
| 16 | 210.0 | 57.0 | 190.0 | 42.0 |
| 17 | 111.0 | 63.0 | 276.0 | 49.0 |
| 18 | 88.0 | 68.0 | 284.0 | 59.0 |
| 19 | 219.0 | 52.0 | 176.0 | 52.0 |
| 20 | 144.0 | 60.0 | 262.0 | 33.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.208 | -1.918 | 1.038 | -1.523 |
| 02 | -2.232 | -2.392 | 1.294 | -1.866 |
| 03 | -2.482 | -2.309 | 1.334 | -3.901 |
| 04 | 1.361 | -2.692 | -4.398 | -3.901 |
| 05 | -1.598 | -3.901 | 1.283 | -1.768 |
| 06 | -2.161 | -0.804 | 1.212 | -2.582 |
| 07 | 1.313 | -3.124 | -1.638 | -3.124 |
| 08 | -0.858 | -0.17 | 0.925 | -1.56 |
| 09 | -1.679 | -0.401 | 0.916 | -0.437 |
| 10 | 0.312 | -0.437 | 0.525 | -1.213 |
| 11 | 0.506 | -0.501 | 0.306 | -0.977 |
| 12 | -0.804 | -0.769 | 1.046 | -1.419 |
| 13 | 0.497 | -0.804 | 0.442 | -1.042 |
| 14 | -0.072 | -0.821 | 0.854 | -1.268 |
| 15 | -0.279 | -0.977 | 0.903 | -0.916 |
| 16 | 0.516 | -0.769 | 0.416 | -1.065 |
| 17 | -0.115 | -0.671 | 0.787 | -0.916 |
| 18 | -0.344 | -0.597 | 0.816 | -0.735 |
| 19 | 0.557 | -0.858 | 0.341 | -0.858 |
| 20 | 0.142 | -0.719 | 0.736 | -1.296 |