Model info
| Transcription factor | ZNF394 (GeneCards) | ||||||||
| Model | ZN394_HUMAN.H11MO.0.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 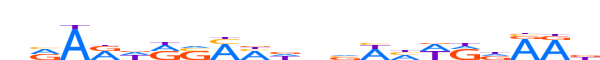 | ||||||||
| LOGO (reverse complement) | 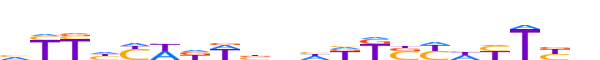 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 20 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nRARdRSMMdnvWhWRdRRd | ||||||||
| Best auROC (human) | 0.802 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 375 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:18832 | ||||||||
| EntrezGene | GeneID:84124 (SSTAR profile) | ||||||||
| UniProt ID | ZN394_HUMAN | ||||||||
| UniProt AC | Q53GI3 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF394 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF394 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 108.0 | 92.0 | 103.0 | 60.0 |
| 02 | 136.0 | 53.0 | 165.0 | 9.0 |
| 03 | 306.0 | 4.0 | 9.0 | 44.0 |
| 04 | 206.0 | 13.0 | 114.0 | 30.0 |
| 05 | 106.0 | 23.0 | 57.0 | 177.0 |
| 06 | 115.0 | 0.0 | 195.0 | 53.0 |
| 07 | 56.0 | 73.0 | 218.0 | 16.0 |
| 08 | 252.0 | 76.0 | 7.0 | 28.0 |
| 09 | 229.0 | 61.0 | 29.0 | 44.0 |
| 10 | 106.0 | 17.0 | 77.0 | 163.0 |
| 11 | 114.0 | 84.0 | 101.0 | 64.0 |
| 12 | 62.0 | 70.0 | 179.0 | 52.0 |
| 13 | 229.0 | 40.0 | 16.0 | 78.0 |
| 14 | 183.0 | 100.0 | 22.0 | 58.0 |
| 15 | 145.0 | 5.0 | 27.0 | 186.0 |
| 16 | 66.0 | 10.0 | 233.0 | 54.0 |
| 17 | 93.0 | 25.0 | 178.0 | 67.0 |
| 18 | 280.0 | 22.0 | 46.0 | 15.0 |
| 19 | 275.0 | 9.0 | 51.0 | 28.0 |
| 20 | 46.0 | 38.0 | 120.0 | 159.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.171 | 0.013 | 0.125 | -0.406 |
| 02 | 0.399 | -0.526 | 0.591 | -2.175 |
| 03 | 1.204 | -2.824 | -2.175 | -0.707 |
| 04 | 0.811 | -1.852 | 0.225 | -1.075 |
| 05 | 0.153 | -1.327 | -0.456 | 0.66 |
| 06 | 0.233 | -4.137 | 0.756 | -0.526 |
| 07 | -0.473 | -0.214 | 0.867 | -1.664 |
| 08 | 1.011 | -0.174 | -2.387 | -1.141 |
| 09 | 0.916 | -0.389 | -1.107 | -0.707 |
| 10 | 0.153 | -1.608 | -0.161 | 0.579 |
| 11 | 0.225 | -0.076 | 0.105 | -0.343 |
| 12 | -0.374 | -0.255 | 0.671 | -0.545 |
| 13 | 0.916 | -0.799 | -1.664 | -0.149 |
| 14 | 0.693 | 0.096 | -1.368 | -0.439 |
| 15 | 0.463 | -2.656 | -1.175 | 0.709 |
| 16 | -0.312 | -2.084 | 0.933 | -0.508 |
| 17 | 0.024 | -1.248 | 0.666 | -0.298 |
| 18 | 1.116 | -1.368 | -0.664 | -1.722 |
| 19 | 1.098 | -2.175 | -0.564 | -1.141 |
| 20 | -0.664 | -0.849 | 0.275 | 0.554 |