Model info
| Transcription factor | Tfap2c | ||||||||
| Model | AP2C_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 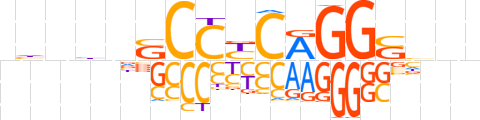 | ||||||||
| LOGO (reverse complement) | 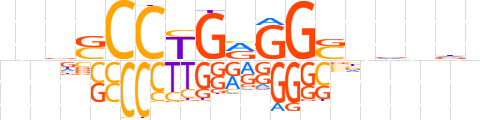 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 17 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nhnbnSCCYCAGGShnn | ||||||||
| Best auROC (human) | 0.954 | ||||||||
| Best auROC (mouse) | 0.962 | ||||||||
| Peak sets in benchmark (human) | 18 | ||||||||
| Peak sets in benchmark (mouse) | 6 | ||||||||
| Aligned words | 493 | ||||||||
| TF family | AP-2 {1.3.1} | ||||||||
| TF subfamily | AP-2gamma {1.3.1.0.3} | ||||||||
| MGI | MGI:106032 | ||||||||
| EntrezGene | GeneID:21420 (SSTAR profile) | ||||||||
| UniProt ID | AP2C_MOUSE | ||||||||
| UniProt AC | Q61312 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Tfap2c expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 31.0 | 35.0 | 18.0 | 18.0 | 34.0 | 82.0 | 12.0 | 54.0 | 33.0 | 59.0 | 14.0 | 17.0 | 8.0 | 43.0 | 11.0 | 23.0 |
| 02 | 41.0 | 24.0 | 35.0 | 6.0 | 74.0 | 74.0 | 22.0 | 49.0 | 13.0 | 7.0 | 19.0 | 16.0 | 13.0 | 37.0 | 35.0 | 27.0 |
| 03 | 19.0 | 37.0 | 36.0 | 49.0 | 24.0 | 50.0 | 9.0 | 59.0 | 15.0 | 39.0 | 26.0 | 31.0 | 7.0 | 32.0 | 19.0 | 40.0 |
| 04 | 3.0 | 15.0 | 32.0 | 15.0 | 62.0 | 30.0 | 30.0 | 36.0 | 13.0 | 33.0 | 29.0 | 15.0 | 38.0 | 25.0 | 82.0 | 34.0 |
| 05 | 5.0 | 20.0 | 87.0 | 4.0 | 7.0 | 26.0 | 70.0 | 0.0 | 5.0 | 93.0 | 72.0 | 3.0 | 4.0 | 25.0 | 67.0 | 4.0 |
| 06 | 0.0 | 21.0 | 0.0 | 0.0 | 0.0 | 164.0 | 0.0 | 0.0 | 0.0 | 293.0 | 1.0 | 2.0 | 0.0 | 11.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 408.0 | 0.0 | 81.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 175.0 | 39.0 | 194.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 51.0 | 13.0 | 18.0 |
| 09 | 1.0 | 1.0 | 0.0 | 0.0 | 14.0 | 210.0 | 0.0 | 2.0 | 0.0 | 52.0 | 0.0 | 0.0 | 15.0 | 197.0 | 0.0 | 0.0 |
| 10 | 29.0 | 0.0 | 1.0 | 0.0 | 334.0 | 15.0 | 108.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 |
| 11 | 8.0 | 0.0 | 355.0 | 2.0 | 1.0 | 0.0 | 14.0 | 0.0 | 3.0 | 0.0 | 106.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 |
| 12 | 1.0 | 0.0 | 11.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 477.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 13 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 11.0 | 227.0 | 247.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 1.0 | 8.0 | 1.0 | 1.0 | 77.0 | 44.0 | 48.0 | 60.0 | 45.0 | 150.0 | 26.0 | 26.0 | 1.0 | 3.0 | 1.0 | 0.0 |
| 15 | 39.0 | 20.0 | 49.0 | 16.0 | 97.0 | 43.0 | 11.0 | 54.0 | 26.0 | 25.0 | 16.0 | 9.0 | 31.0 | 19.0 | 23.0 | 14.0 |
| 16 | 31.0 | 46.0 | 64.0 | 52.0 | 12.0 | 36.0 | 17.0 | 42.0 | 10.0 | 41.0 | 30.0 | 18.0 | 10.0 | 22.0 | 37.0 | 24.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.008 | 0.128 | -0.527 | -0.527 | 0.099 | 0.973 | -0.922 | 0.558 | 0.07 | 0.646 | -0.772 | -0.583 | -1.312 | 0.332 | -1.006 | -0.286 |
| 02 | 0.285 | -0.244 | 0.128 | -1.584 | 0.871 | 0.871 | -0.33 | 0.461 | -0.844 | -1.439 | -0.474 | -0.642 | -0.844 | 0.183 | 0.128 | -0.128 |
| 03 | -0.474 | 0.183 | 0.156 | 0.461 | -0.244 | 0.481 | -1.199 | 0.646 | -0.705 | 0.235 | -0.166 | 0.008 | -1.439 | 0.039 | -0.474 | 0.26 |
| 04 | -2.218 | -0.705 | 0.039 | -0.705 | 0.695 | -0.024 | -0.024 | 0.156 | -0.844 | 0.07 | -0.058 | -0.705 | 0.209 | -0.204 | 0.973 | 0.099 |
| 05 | -1.754 | -0.423 | 1.032 | -1.96 | -1.439 | -0.166 | 0.816 | -4.387 | -1.754 | 1.098 | 0.844 | -2.218 | -1.96 | -0.204 | 0.772 | -1.96 |
| 06 | -4.387 | -0.376 | -4.387 | -4.387 | -4.387 | 1.664 | -4.387 | -4.387 | -4.387 | 2.243 | -3.111 | -2.568 | -4.387 | -1.006 | -4.387 | -4.387 |
| 07 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | 2.574 | -4.387 | 0.961 | -4.387 | -3.111 | -4.387 | -4.387 | -4.387 | -3.111 | -4.387 | -3.111 |
| 08 | -4.387 | -4.387 | -4.387 | -4.387 | -2.568 | 1.729 | 0.235 | 1.831 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | 0.501 | -0.844 | -0.527 |
| 09 | -3.111 | -3.111 | -4.387 | -4.387 | -0.772 | 1.911 | -4.387 | -2.568 | -4.387 | 0.52 | -4.387 | -4.387 | -0.705 | 1.847 | -4.387 | -4.387 |
| 10 | -0.058 | -4.387 | -3.111 | -4.387 | 2.374 | -0.705 | 1.247 | -2.218 | -4.387 | -4.387 | -4.387 | -4.387 | -2.568 | -4.387 | -4.387 | -4.387 |
| 11 | -1.312 | -4.387 | 2.435 | -2.568 | -3.111 | -4.387 | -0.772 | -4.387 | -2.218 | -4.387 | 1.229 | -4.387 | -4.387 | -4.387 | -2.218 | -4.387 |
| 12 | -3.111 | -4.387 | -1.006 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -3.111 | 2.73 | -4.387 | -4.387 | -4.387 | -2.568 | -4.387 |
| 13 | -4.387 | -3.111 | -4.387 | -4.387 | -4.387 | -3.111 | -4.387 | -4.387 | -1.006 | 1.988 | 2.073 | -1.754 | -4.387 | -4.387 | -4.387 | -4.387 |
| 14 | -3.111 | -1.312 | -3.111 | -3.111 | 0.91 | 0.355 | 0.441 | 0.662 | 0.377 | 1.575 | -0.166 | -0.166 | -3.111 | -2.218 | -3.111 | -4.387 |
| 15 | 0.235 | -0.423 | 0.461 | -0.642 | 1.14 | 0.332 | -1.006 | 0.558 | -0.166 | -0.204 | -0.642 | -1.199 | 0.008 | -0.474 | -0.286 | -0.772 |
| 16 | 0.008 | 0.399 | 0.727 | 0.52 | -0.922 | 0.156 | -0.583 | 0.308 | -1.098 | 0.285 | -0.024 | -0.527 | -1.098 | -0.33 | 0.183 | -0.244 |