Model info
| Transcription factor | ARNTL (GeneCards) | ||||||||
| Model | BMAL1_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 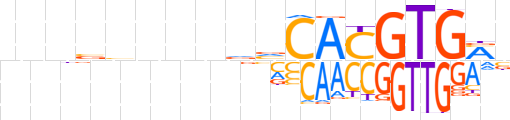 | ||||||||
| LOGO (reverse complement) | 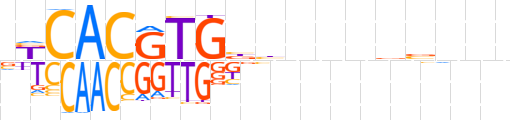 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bnbvnnnvdvCACGTGMb | ||||||||
| Best auROC (human) | 0.881 | ||||||||
| Best auROC (mouse) | 0.868 | ||||||||
| Peak sets in benchmark (human) | 18 | ||||||||
| Peak sets in benchmark (mouse) | 15 | ||||||||
| Aligned words | 503 | ||||||||
| TF family | PAS domain factors {1.2.5} | ||||||||
| TF subfamily | Arnt-like factors {1.2.5.2} | ||||||||
| HGNC | HGNC:701 | ||||||||
| EntrezGene | GeneID:406 (SSTAR profile) | ||||||||
| UniProt ID | BMAL1_HUMAN | ||||||||
| UniProt AC | O00327 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ARNTL expression | ||||||||
| ReMap ChIP-seq dataset list | ARNTL datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 14.0 | 10.0 | 9.0 | 26.0 | 65.0 | 57.0 | 55.0 | 27.0 | 25.0 | 48.0 | 18.0 | 51.0 | 13.0 | 12.0 | 46.0 | 19.0 |
| 02 | 20.0 | 19.0 | 30.0 | 48.0 | 21.0 | 24.0 | 40.0 | 42.0 | 19.0 | 32.0 | 26.0 | 51.0 | 18.0 | 7.0 | 67.0 | 31.0 |
| 03 | 32.0 | 14.0 | 28.0 | 4.0 | 15.0 | 30.0 | 25.0 | 12.0 | 27.0 | 57.0 | 64.0 | 15.0 | 8.0 | 41.0 | 103.0 | 20.0 |
| 04 | 14.0 | 35.0 | 20.0 | 13.0 | 27.0 | 64.0 | 17.0 | 34.0 | 43.0 | 76.0 | 34.0 | 67.0 | 2.0 | 21.0 | 15.0 | 13.0 |
| 05 | 20.0 | 22.0 | 29.0 | 15.0 | 48.0 | 68.0 | 39.0 | 41.0 | 19.0 | 42.0 | 11.0 | 14.0 | 23.0 | 27.0 | 65.0 | 12.0 |
| 06 | 19.0 | 25.0 | 50.0 | 16.0 | 43.0 | 42.0 | 31.0 | 43.0 | 27.0 | 39.0 | 53.0 | 25.0 | 8.0 | 35.0 | 29.0 | 10.0 |
| 07 | 19.0 | 14.0 | 40.0 | 24.0 | 57.0 | 24.0 | 40.0 | 20.0 | 38.0 | 35.0 | 63.0 | 27.0 | 12.0 | 18.0 | 50.0 | 14.0 |
| 08 | 29.0 | 21.0 | 58.0 | 18.0 | 31.0 | 12.0 | 30.0 | 18.0 | 51.0 | 28.0 | 86.0 | 28.0 | 11.0 | 9.0 | 41.0 | 24.0 |
| 09 | 51.0 | 33.0 | 31.0 | 7.0 | 25.0 | 35.0 | 10.0 | 0.0 | 47.0 | 114.0 | 38.0 | 16.0 | 23.0 | 31.0 | 30.0 | 4.0 |
| 10 | 10.0 | 134.0 | 2.0 | 0.0 | 9.0 | 196.0 | 5.0 | 3.0 | 8.0 | 97.0 | 2.0 | 2.0 | 1.0 | 26.0 | 0.0 | 0.0 |
| 11 | 20.0 | 0.0 | 7.0 | 1.0 | 422.0 | 6.0 | 18.0 | 7.0 | 8.0 | 0.0 | 1.0 | 0.0 | 3.0 | 2.0 | 0.0 | 0.0 |
| 12 | 7.0 | 365.0 | 4.0 | 77.0 | 3.0 | 2.0 | 0.0 | 3.0 | 3.0 | 19.0 | 0.0 | 4.0 | 0.0 | 7.0 | 0.0 | 1.0 |
| 13 | 0.0 | 0.0 | 13.0 | 0.0 | 6.0 | 6.0 | 379.0 | 2.0 | 0.0 | 0.0 | 4.0 | 0.0 | 3.0 | 2.0 | 75.0 | 5.0 |
| 14 | 0.0 | 0.0 | 0.0 | 9.0 | 0.0 | 1.0 | 0.0 | 7.0 | 7.0 | 2.0 | 1.0 | 461.0 | 0.0 | 0.0 | 0.0 | 7.0 |
| 15 | 0.0 | 0.0 | 6.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 8.0 | 1.0 | 464.0 | 11.0 |
| 16 | 2.0 | 1.0 | 5.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 328.0 | 65.0 | 39.0 | 42.0 | 7.0 | 0.0 | 2.0 | 3.0 |
| 17 | 28.0 | 149.0 | 118.0 | 42.0 | 24.0 | 14.0 | 15.0 | 14.0 | 5.0 | 7.0 | 22.0 | 12.0 | 7.0 | 10.0 | 13.0 | 15.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.778 | -1.104 | -1.205 | -0.172 | 0.736 | 0.605 | 0.57 | -0.134 | -0.21 | 0.435 | -0.533 | 0.495 | -0.85 | -0.928 | 0.393 | -0.48 |
| 02 | -0.429 | -0.48 | -0.03 | 0.435 | -0.382 | -0.25 | 0.254 | 0.302 | -0.48 | 0.033 | -0.172 | 0.495 | -0.533 | -1.445 | 0.766 | 0.002 |
| 03 | 0.033 | -0.778 | -0.098 | -1.966 | -0.711 | -0.03 | -0.21 | -0.928 | -0.134 | 0.605 | 0.72 | -0.711 | -1.318 | 0.279 | 1.194 | -0.429 |
| 04 | -0.778 | 0.122 | -0.429 | -0.85 | -0.134 | 0.72 | -0.589 | 0.093 | 0.326 | 0.891 | 0.093 | 0.766 | -2.574 | -0.382 | -0.711 | -0.85 |
| 05 | -0.429 | -0.336 | -0.064 | -0.711 | 0.435 | 0.781 | 0.229 | 0.279 | -0.48 | 0.302 | -1.012 | -0.778 | -0.292 | -0.134 | 0.736 | -0.928 |
| 06 | -0.48 | -0.21 | 0.475 | -0.648 | 0.326 | 0.302 | 0.002 | 0.326 | -0.134 | 0.229 | 0.533 | -0.21 | -1.318 | 0.122 | -0.064 | -1.104 |
| 07 | -0.48 | -0.778 | 0.254 | -0.25 | 0.605 | -0.25 | 0.254 | -0.429 | 0.203 | 0.122 | 0.705 | -0.134 | -0.928 | -0.533 | 0.475 | -0.778 |
| 08 | -0.064 | -0.382 | 0.623 | -0.533 | 0.002 | -0.928 | -0.03 | -0.533 | 0.495 | -0.098 | 1.014 | -0.098 | -1.012 | -1.205 | 0.279 | -0.25 |
| 09 | 0.495 | 0.064 | 0.002 | -1.445 | -0.21 | 0.122 | -1.104 | -4.392 | 0.414 | 1.295 | 0.203 | -0.648 | -0.292 | 0.002 | -0.03 | -1.966 |
| 10 | -1.104 | 1.456 | -2.574 | -4.392 | -1.205 | 1.836 | -1.76 | -2.224 | -1.318 | 1.134 | -2.574 | -2.574 | -3.117 | -0.172 | -4.392 | -4.392 |
| 11 | -0.429 | -4.392 | -1.445 | -3.117 | 2.601 | -1.59 | -0.533 | -1.445 | -1.318 | -4.392 | -3.117 | -4.392 | -2.224 | -2.574 | -4.392 | -4.392 |
| 12 | -1.445 | 2.457 | -1.966 | 0.904 | -2.224 | -2.574 | -4.392 | -2.224 | -2.224 | -0.48 | -4.392 | -1.966 | -4.392 | -1.445 | -4.392 | -3.117 |
| 13 | -4.392 | -4.392 | -0.85 | -4.392 | -1.59 | -1.59 | 2.494 | -2.574 | -4.392 | -4.392 | -1.966 | -4.392 | -2.224 | -2.574 | 0.878 | -1.76 |
| 14 | -4.392 | -4.392 | -4.392 | -1.205 | -4.392 | -3.117 | -4.392 | -1.445 | -1.445 | -2.574 | -3.117 | 2.69 | -4.392 | -4.392 | -4.392 | -1.445 |
| 15 | -4.392 | -4.392 | -1.59 | -3.117 | -4.392 | -4.392 | -2.224 | -4.392 | -4.392 | -4.392 | -3.117 | -4.392 | -1.318 | -3.117 | 2.696 | -1.012 |
| 16 | -2.574 | -3.117 | -1.76 | -4.392 | -4.392 | -3.117 | -4.392 | -4.392 | 2.35 | 0.736 | 0.229 | 0.302 | -1.445 | -4.392 | -2.574 | -2.224 |
| 17 | -0.098 | 1.562 | 1.33 | 0.302 | -0.25 | -0.778 | -0.711 | -0.778 | -1.76 | -1.445 | -0.336 | -0.928 | -1.445 | -1.104 | -0.85 | -0.711 |