Model info
| Transcription factor | Cebpa | ||||||||
| Model | CEBPA_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 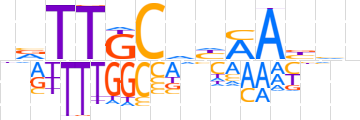 | ||||||||
| LOGO (reverse complement) | 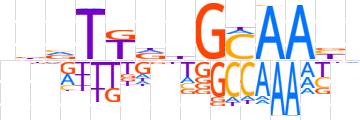 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 13 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvTTGCdhMAbhn | ||||||||
| Best auROC (human) | 0.987 | ||||||||
| Best auROC (mouse) | 0.992 | ||||||||
| Peak sets in benchmark (human) | 18 | ||||||||
| Peak sets in benchmark (mouse) | 153 | ||||||||
| Aligned words | 411 | ||||||||
| TF family | C/EBP-related {1.1.8} | ||||||||
| TF subfamily | C/EBP {1.1.8.1} | ||||||||
| MGI | MGI:99480 | ||||||||
| EntrezGene | GeneID:12606 (SSTAR profile) | ||||||||
| UniProt ID | CEBPA_MOUSE | ||||||||
| UniProt AC | P53566 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Cebpa expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 31.0 | 5.0 | 45.0 | 1.0 | 51.0 | 18.0 | 14.0 | 2.0 | 47.0 | 18.0 | 41.0 | 5.0 | 59.0 | 34.0 | 36.0 | 1.0 |
| 02 | 0.0 | 1.0 | 0.0 | 187.0 | 0.0 | 1.0 | 0.0 | 74.0 | 0.0 | 1.0 | 3.0 | 132.0 | 0.0 | 2.0 | 0.0 | 7.0 |
| 03 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 10.0 | 390.0 |
| 04 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 4.0 | 4.0 | 35.0 | 1.0 | 319.0 | 42.0 |
| 05 | 0.0 | 36.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 321.0 | 2.0 | 1.0 | 1.0 | 45.0 | 0.0 | 0.0 |
| 06 | 0.0 | 1.0 | 0.0 | 0.0 | 194.0 | 62.0 | 73.0 | 74.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 |
| 07 | 40.0 | 104.0 | 9.0 | 42.0 | 16.0 | 29.0 | 0.0 | 19.0 | 19.0 | 34.0 | 1.0 | 20.0 | 7.0 | 35.0 | 0.0 | 33.0 |
| 08 | 28.0 | 53.0 | 1.0 | 0.0 | 188.0 | 13.0 | 0.0 | 1.0 | 2.0 | 7.0 | 1.0 | 0.0 | 44.0 | 69.0 | 1.0 | 0.0 |
| 09 | 253.0 | 1.0 | 3.0 | 5.0 | 139.0 | 2.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 10 | 18.0 | 160.0 | 67.0 | 148.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 | 2.0 | 1.0 | 1.0 | 1.0 | 4.0 | 1.0 | 2.0 |
| 11 | 6.0 | 5.0 | 2.0 | 7.0 | 64.0 | 52.0 | 12.0 | 38.0 | 33.0 | 19.0 | 7.0 | 10.0 | 39.0 | 62.0 | 27.0 | 25.0 |
| 12 | 23.0 | 35.0 | 50.0 | 34.0 | 31.0 | 44.0 | 11.0 | 52.0 | 14.0 | 11.0 | 10.0 | 13.0 | 9.0 | 33.0 | 17.0 | 21.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.193 | -1.571 | 0.562 | -2.934 | 0.686 | -0.342 | -0.588 | -2.388 | 0.605 | -0.342 | 0.469 | -1.571 | 0.831 | 0.284 | 0.341 | -2.934 |
| 02 | -4.232 | -2.934 | -4.232 | 1.98 | -4.232 | -2.934 | -4.232 | 1.056 | -4.232 | -2.934 | -2.037 | 1.632 | -4.232 | -2.388 | -4.232 | -1.255 |
| 03 | -4.232 | -4.232 | -4.232 | -4.232 | -4.232 | -4.232 | -4.232 | -1.571 | -2.934 | -4.232 | -4.232 | -2.388 | -4.232 | -4.232 | -0.914 | 2.714 |
| 04 | -4.232 | -4.232 | -2.934 | -4.232 | -4.232 | -4.232 | -4.232 | -4.232 | -2.388 | -4.232 | -1.777 | -1.777 | 0.313 | -2.934 | 2.513 | 0.493 |
| 05 | -4.232 | 0.341 | -4.232 | -2.934 | -4.232 | -2.934 | -4.232 | -4.232 | -4.232 | 2.519 | -2.388 | -2.934 | -2.934 | 0.562 | -4.232 | -4.232 |
| 06 | -4.232 | -2.934 | -4.232 | -4.232 | 2.016 | 0.88 | 1.042 | 1.056 | -2.934 | -2.934 | -4.232 | -4.232 | -4.232 | -4.232 | -2.934 | -2.934 |
| 07 | 0.445 | 1.395 | -1.015 | 0.493 | -0.458 | 0.127 | -4.232 | -0.289 | -0.289 | 0.284 | -2.934 | -0.239 | -1.255 | 0.313 | -4.232 | 0.255 |
| 08 | 0.092 | 0.724 | -2.934 | -4.232 | 1.985 | -0.66 | -4.232 | -2.934 | -2.388 | -1.255 | -2.934 | -4.232 | 0.539 | 0.986 | -2.934 | -4.232 |
| 09 | 2.282 | -2.934 | -2.037 | -1.571 | 1.684 | -2.388 | -4.232 | -2.934 | -2.934 | -4.232 | -4.232 | -2.388 | -4.232 | -4.232 | -2.934 | -4.232 |
| 10 | -0.342 | 1.824 | 0.957 | 1.746 | -2.934 | -4.232 | -4.232 | -2.388 | -4.232 | -2.388 | -2.934 | -2.934 | -2.934 | -1.777 | -2.934 | -2.388 |
| 11 | -1.401 | -1.571 | -2.388 | -1.255 | 0.911 | 0.705 | -0.738 | 0.394 | 0.255 | -0.289 | -1.255 | -0.914 | 0.42 | 0.88 | 0.056 | -0.02 |
| 12 | -0.102 | 0.313 | 0.666 | 0.284 | 0.193 | 0.539 | -0.822 | 0.705 | -0.588 | -0.822 | -0.914 | -0.66 | -1.015 | 0.255 | -0.398 | -0.191 |