Model info
| Transcription factor | NR2F1 (GeneCards) | ||||||||
| Model | COT1_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 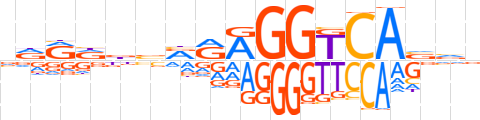 | ||||||||
| LOGO (reverse complement) | 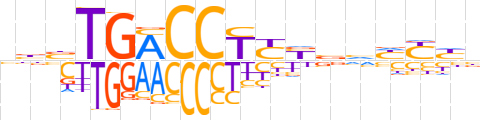 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 17 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vdRdbbRRAGGTCAvvv | ||||||||
| Best auROC (human) | 0.868 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 489 | ||||||||
| TF family | RXR-related receptors (NR2) {2.1.3} | ||||||||
| TF subfamily | COUP-like receptors (NR2F) {2.1.3.5} | ||||||||
| HGNC | HGNC:7975 | ||||||||
| EntrezGene | GeneID:7025 (SSTAR profile) | ||||||||
| UniProt ID | COT1_HUMAN | ||||||||
| UniProt AC | P10589 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | NR2F1 expression | ||||||||
| ReMap ChIP-seq dataset list | NR2F1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 35.0 | 5.0 | 78.0 | 6.0 | 56.0 | 9.0 | 17.0 | 17.0 | 72.0 | 9.0 | 90.0 | 15.0 | 12.0 | 1.0 | 48.0 | 12.0 |
| 02 | 32.0 | 6.0 | 129.0 | 8.0 | 11.0 | 5.0 | 7.0 | 1.0 | 48.0 | 22.0 | 152.0 | 11.0 | 7.0 | 4.0 | 32.0 | 7.0 |
| 03 | 18.0 | 13.0 | 55.0 | 12.0 | 10.0 | 8.0 | 11.0 | 8.0 | 45.0 | 30.0 | 207.0 | 38.0 | 0.0 | 9.0 | 14.0 | 4.0 |
| 04 | 10.0 | 15.0 | 32.0 | 16.0 | 13.0 | 14.0 | 10.0 | 23.0 | 18.0 | 42.0 | 94.0 | 133.0 | 4.0 | 16.0 | 28.0 | 14.0 |
| 05 | 10.0 | 10.0 | 22.0 | 3.0 | 30.0 | 33.0 | 3.0 | 21.0 | 22.0 | 78.0 | 36.0 | 28.0 | 7.0 | 110.0 | 47.0 | 22.0 |
| 06 | 37.0 | 10.0 | 17.0 | 5.0 | 168.0 | 21.0 | 23.0 | 19.0 | 55.0 | 19.0 | 24.0 | 10.0 | 28.0 | 10.0 | 29.0 | 7.0 |
| 07 | 100.0 | 11.0 | 172.0 | 5.0 | 32.0 | 0.0 | 23.0 | 5.0 | 35.0 | 3.0 | 51.0 | 4.0 | 13.0 | 0.0 | 28.0 | 0.0 |
| 08 | 139.0 | 0.0 | 41.0 | 0.0 | 12.0 | 0.0 | 1.0 | 1.0 | 173.0 | 2.0 | 99.0 | 0.0 | 10.0 | 0.0 | 4.0 | 0.0 |
| 09 | 5.0 | 0.0 | 324.0 | 5.0 | 0.0 | 0.0 | 2.0 | 0.0 | 1.0 | 0.0 | 143.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 10 | 0.0 | 0.0 | 6.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 2.0 | 461.0 | 4.0 | 0.0 | 0.0 | 6.0 | 0.0 |
| 11 | 0.0 | 0.0 | 2.0 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 26.0 | 86.0 | 361.0 | 0.0 | 0.0 | 0.0 | 4.0 |
| 12 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 24.0 | 1.0 | 0.0 | 3.0 | 84.0 | 1.0 | 0.0 | 3.0 | 347.0 | 11.0 | 7.0 |
| 13 | 7.0 | 0.0 | 0.0 | 0.0 | 438.0 | 2.0 | 11.0 | 4.0 | 13.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 |
| 14 | 78.0 | 95.0 | 250.0 | 42.0 | 0.0 | 0.0 | 1.0 | 1.0 | 4.0 | 6.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.0 |
| 15 | 31.0 | 7.0 | 40.0 | 4.0 | 38.0 | 20.0 | 28.0 | 16.0 | 92.0 | 66.0 | 81.0 | 15.0 | 7.0 | 8.0 | 24.0 | 5.0 |
| 16 | 24.0 | 25.0 | 96.0 | 23.0 | 39.0 | 28.0 | 20.0 | 14.0 | 32.0 | 33.0 | 81.0 | 27.0 | 4.0 | 5.0 | 23.0 | 8.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.148 | -1.734 | 0.944 | -1.564 | 0.614 | -1.179 | -0.562 | -0.562 | 0.864 | -1.179 | 1.086 | -0.685 | -0.902 | -3.092 | 0.461 | -0.902 |
| 02 | 0.06 | -1.564 | 1.445 | -1.292 | -0.986 | -1.734 | -1.418 | -3.092 | 0.461 | -0.31 | 1.608 | -0.986 | -1.418 | -1.94 | 0.06 | -1.418 |
| 03 | -0.506 | -0.824 | 0.596 | -0.902 | -1.078 | -1.292 | -0.986 | -1.292 | 0.397 | -0.004 | 1.916 | 0.23 | -4.37 | -1.179 | -0.752 | -1.94 |
| 04 | -1.078 | -0.685 | 0.06 | -0.622 | -0.824 | -0.752 | -1.078 | -0.266 | -0.506 | 0.329 | 1.129 | 1.475 | -1.94 | -0.622 | -0.072 | -0.752 |
| 05 | -1.078 | -1.078 | -0.31 | -2.198 | -0.004 | 0.09 | -2.198 | -0.355 | -0.31 | 0.944 | 0.176 | -0.072 | -1.418 | 1.286 | 0.44 | -0.31 |
| 06 | 0.203 | -1.078 | -0.562 | -1.734 | 1.708 | -0.355 | -0.266 | -0.454 | 0.596 | -0.454 | -0.224 | -1.078 | -0.072 | -1.078 | -0.038 | -1.418 |
| 07 | 1.191 | -0.986 | 1.732 | -1.734 | 0.06 | -4.37 | -0.266 | -1.734 | 0.148 | -2.198 | 0.521 | -1.94 | -0.824 | -4.37 | -0.072 | -4.37 |
| 08 | 1.519 | -4.37 | 0.305 | -4.37 | -0.902 | -4.37 | -3.092 | -3.092 | 1.737 | -2.548 | 1.181 | -4.37 | -1.078 | -4.37 | -1.94 | -4.37 |
| 09 | -1.734 | -4.37 | 2.364 | -1.734 | -4.37 | -4.37 | -2.548 | -4.37 | -3.092 | -4.37 | 1.547 | -3.092 | -4.37 | -4.37 | -3.092 | -4.37 |
| 10 | -4.37 | -4.37 | -1.564 | -4.37 | -4.37 | -4.37 | -4.37 | -4.37 | -2.198 | -2.548 | 2.716 | -1.94 | -4.37 | -4.37 | -1.564 | -4.37 |
| 11 | -4.37 | -4.37 | -2.548 | -3.092 | -4.37 | -4.37 | -4.37 | -2.548 | -4.37 | -0.145 | 1.041 | 2.472 | -4.37 | -4.37 | -4.37 | -1.94 |
| 12 | -4.37 | -4.37 | -4.37 | -4.37 | -3.092 | -0.224 | -3.092 | -4.37 | -2.198 | 1.017 | -3.092 | -4.37 | -2.198 | 2.432 | -0.986 | -1.418 |
| 13 | -1.418 | -4.37 | -4.37 | -4.37 | 2.665 | -2.548 | -0.986 | -1.94 | -0.824 | -4.37 | -4.37 | -4.37 | -1.418 | -4.37 | -4.37 | -4.37 |
| 14 | 0.944 | 1.14 | 2.105 | 0.329 | -4.37 | -4.37 | -3.092 | -3.092 | -1.94 | -1.564 | -4.37 | -3.092 | -4.37 | -3.092 | -2.198 | -4.37 |
| 15 | 0.028 | -1.418 | 0.28 | -1.94 | 0.23 | -0.403 | -0.072 | -0.622 | 1.108 | 0.777 | 0.981 | -0.685 | -1.418 | -1.292 | -0.224 | -1.734 |
| 16 | -0.224 | -0.184 | 1.15 | -0.266 | 0.255 | -0.072 | -0.403 | -0.752 | 0.06 | 0.09 | 0.981 | -0.108 | -1.94 | -1.734 | -0.266 | -1.292 |