Model info
| Transcription factor | FOXP2 (GeneCards) | ||||||||
| Model | FOXP2_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 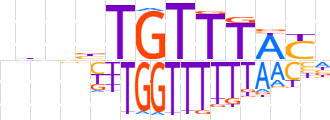 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbbbTGTTTWYn | ||||||||
| Best auROC (human) | 0.97 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 509 | ||||||||
| TF family | Forkhead box (FOX) factors {3.3.1} | ||||||||
| TF subfamily | FOXP {3.3.1.16} | ||||||||
| HGNC | HGNC:13875 | ||||||||
| EntrezGene | GeneID:93986 (SSTAR profile) | ||||||||
| UniProt ID | FOXP2_HUMAN | ||||||||
| UniProt AC | O15409 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | FOXP2 expression | ||||||||
| ReMap ChIP-seq dataset list | FOXP2 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 17.0 | 15.0 | 27.0 | 34.0 | 25.0 | 22.0 | 31.0 | 31.0 | 16.0 | 35.0 | 32.0 | 22.0 | 16.0 | 24.0 | 40.0 | 100.0 |
| 02 | 8.0 | 17.0 | 27.0 | 22.0 | 20.0 | 18.0 | 11.0 | 47.0 | 16.0 | 28.0 | 45.0 | 41.0 | 8.0 | 90.0 | 55.0 | 34.0 |
| 03 | 6.0 | 14.0 | 19.0 | 13.0 | 1.0 | 102.0 | 13.0 | 37.0 | 21.0 | 49.0 | 21.0 | 47.0 | 5.0 | 31.0 | 49.0 | 59.0 |
| 04 | 0.0 | 0.0 | 0.0 | 33.0 | 2.0 | 1.0 | 3.0 | 190.0 | 0.0 | 0.0 | 0.0 | 102.0 | 1.0 | 1.0 | 2.0 | 152.0 |
| 05 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 26.0 | 0.0 | 451.0 | 0.0 |
| 06 | 0.0 | 1.0 | 0.0 | 25.0 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 3.0 | 1.0 | 454.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 2.0 | 34.0 | 444.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | 4.0 | 29.0 | 0.0 | 1.0 | 60.0 | 390.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 38.0 | 12.0 | 0.0 | 15.0 | 353.0 | 19.0 | 30.0 | 17.0 |
| 10 | 1.0 | 293.0 | 8.0 | 90.0 | 1.0 | 22.0 | 0.0 | 9.0 | 3.0 | 0.0 | 3.0 | 24.0 | 0.0 | 14.0 | 5.0 | 14.0 |
| 11 | 1.0 | 0.0 | 4.0 | 0.0 | 137.0 | 87.0 | 16.0 | 89.0 | 0.0 | 6.0 | 7.0 | 3.0 | 12.0 | 33.0 | 47.0 | 45.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.573 | -0.695 | -0.118 | 0.109 | -0.194 | -0.32 | 0.018 | 0.018 | -0.632 | 0.138 | 0.049 | -0.32 | -0.632 | -0.234 | 0.27 | 1.181 |
| 02 | -1.302 | -0.573 | -0.118 | -0.32 | -0.413 | -0.517 | -0.996 | 0.43 | -0.632 | -0.082 | 0.387 | 0.295 | -1.302 | 1.076 | 0.586 | 0.109 |
| 03 | -1.574 | -0.762 | -0.464 | -0.834 | -3.101 | 1.2 | -0.834 | 0.193 | -0.366 | 0.471 | -0.366 | 0.43 | -1.744 | 0.018 | 0.471 | 0.656 |
| 04 | -4.378 | -4.378 | -4.378 | 0.08 | -2.558 | -3.101 | -2.208 | 1.821 | -4.378 | -4.378 | -4.378 | 1.2 | -3.101 | -3.101 | -2.558 | 1.598 |
| 05 | -4.378 | -4.378 | -2.208 | -4.378 | -4.378 | -3.101 | -3.101 | -4.378 | -4.378 | -4.378 | -1.744 | -4.378 | -0.155 | -4.378 | 2.684 | -4.378 |
| 06 | -4.378 | -3.101 | -4.378 | -0.194 | -4.378 | -4.378 | -4.378 | -3.101 | -2.558 | -2.208 | -3.101 | 2.691 | -4.378 | -4.378 | -4.378 | -4.378 |
| 07 | -4.378 | -4.378 | -4.378 | -2.558 | -4.378 | -4.378 | -4.378 | -1.95 | -4.378 | -4.378 | -4.378 | -3.101 | -4.378 | -2.558 | 0.109 | 2.668 |
| 08 | -4.378 | -4.378 | -4.378 | -4.378 | -4.378 | -3.101 | -3.101 | -4.378 | -4.378 | -3.101 | -1.95 | -0.048 | -4.378 | -3.101 | 0.672 | 2.539 |
| 09 | -4.378 | -4.378 | -4.378 | -4.378 | -3.101 | -3.101 | -4.378 | -3.101 | 0.219 | -0.912 | -4.378 | -0.695 | 2.439 | -0.464 | -0.014 | -0.573 |
| 10 | -3.101 | 2.253 | -1.302 | 1.076 | -3.101 | -0.32 | -4.378 | -1.189 | -2.208 | -4.378 | -2.208 | -0.234 | -4.378 | -0.762 | -1.744 | -0.762 |
| 11 | -3.101 | -4.378 | -1.95 | -4.378 | 1.494 | 1.042 | -0.632 | 1.065 | -4.378 | -1.574 | -1.429 | -2.208 | -0.912 | 0.08 | 0.43 | 0.387 |