Model info
| Transcription factor | NR3C1 (GeneCards) | ||||||||
| Model | GCR_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 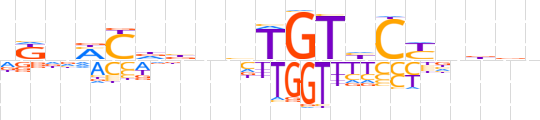 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dvhnRGdACAbnvhGKnMh | ||||||||
| Best auROC (human) | 0.985 | ||||||||
| Best auROC (mouse) | 0.992 | ||||||||
| Peak sets in benchmark (human) | 143 | ||||||||
| Peak sets in benchmark (mouse) | 119 | ||||||||
| Aligned words | 466 | ||||||||
| TF family | Steroid hormone receptors (NR3) {2.1.1} | ||||||||
| TF subfamily | GR-like receptors (NR3C) {2.1.1.1} | ||||||||
| HGNC | HGNC:7978 | ||||||||
| EntrezGene | GeneID:2908 (SSTAR profile) | ||||||||
| UniProt ID | GCR_HUMAN | ||||||||
| UniProt AC | P04150 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | NR3C1 expression | ||||||||
| ReMap ChIP-seq dataset list | NR3C1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 17.0 | 36.0 | 33.0 | 13.0 | 23.0 | 39.0 | 5.0 | 14.0 | 37.0 | 69.0 | 47.0 | 27.0 | 4.0 | 36.0 | 22.0 | 20.0 |
| 02 | 35.0 | 27.0 | 12.0 | 7.0 | 77.0 | 60.0 | 3.0 | 40.0 | 41.0 | 36.0 | 21.0 | 9.0 | 19.0 | 21.0 | 21.0 | 13.0 |
| 03 | 31.0 | 63.0 | 64.0 | 14.0 | 54.0 | 39.0 | 9.0 | 42.0 | 21.0 | 16.0 | 9.0 | 11.0 | 11.0 | 26.0 | 24.0 | 8.0 |
| 04 | 38.0 | 3.0 | 74.0 | 2.0 | 98.0 | 1.0 | 21.0 | 24.0 | 40.0 | 2.0 | 57.0 | 7.0 | 15.0 | 1.0 | 56.0 | 3.0 |
| 05 | 9.0 | 1.0 | 177.0 | 4.0 | 2.0 | 0.0 | 3.0 | 2.0 | 13.0 | 1.0 | 186.0 | 8.0 | 0.0 | 0.0 | 35.0 | 1.0 |
| 06 | 10.0 | 3.0 | 8.0 | 3.0 | 1.0 | 0.0 | 0.0 | 1.0 | 205.0 | 45.0 | 102.0 | 49.0 | 3.0 | 4.0 | 1.0 | 7.0 |
| 07 | 201.0 | 0.0 | 16.0 | 2.0 | 50.0 | 2.0 | 0.0 | 0.0 | 105.0 | 2.0 | 3.0 | 1.0 | 54.0 | 0.0 | 3.0 | 3.0 |
| 08 | 0.0 | 387.0 | 13.0 | 10.0 | 0.0 | 4.0 | 0.0 | 0.0 | 1.0 | 21.0 | 0.0 | 0.0 | 0.0 | 6.0 | 0.0 | 0.0 |
| 09 | 0.0 | 1.0 | 0.0 | 0.0 | 362.0 | 6.0 | 19.0 | 31.0 | 8.0 | 0.0 | 2.0 | 3.0 | 3.0 | 1.0 | 6.0 | 0.0 |
| 10 | 65.0 | 76.0 | 155.0 | 77.0 | 1.0 | 1.0 | 1.0 | 5.0 | 0.0 | 6.0 | 14.0 | 7.0 | 1.0 | 9.0 | 14.0 | 10.0 |
| 11 | 28.0 | 8.0 | 25.0 | 6.0 | 32.0 | 23.0 | 13.0 | 24.0 | 58.0 | 47.0 | 52.0 | 27.0 | 18.0 | 32.0 | 24.0 | 25.0 |
| 12 | 62.0 | 37.0 | 28.0 | 9.0 | 45.0 | 41.0 | 13.0 | 11.0 | 38.0 | 33.0 | 34.0 | 9.0 | 7.0 | 45.0 | 20.0 | 10.0 |
| 13 | 21.0 | 70.0 | 10.0 | 51.0 | 47.0 | 29.0 | 1.0 | 79.0 | 31.0 | 19.0 | 12.0 | 33.0 | 6.0 | 9.0 | 1.0 | 23.0 |
| 14 | 5.0 | 4.0 | 95.0 | 1.0 | 47.0 | 10.0 | 68.0 | 2.0 | 3.0 | 2.0 | 18.0 | 1.0 | 7.0 | 7.0 | 171.0 | 1.0 |
| 15 | 8.0 | 5.0 | 16.0 | 33.0 | 2.0 | 2.0 | 2.0 | 17.0 | 39.0 | 39.0 | 41.0 | 233.0 | 1.0 | 1.0 | 3.0 | 0.0 |
| 16 | 11.0 | 8.0 | 17.0 | 14.0 | 14.0 | 6.0 | 2.0 | 25.0 | 24.0 | 15.0 | 7.0 | 16.0 | 52.0 | 90.0 | 56.0 | 85.0 |
| 17 | 21.0 | 66.0 | 7.0 | 7.0 | 14.0 | 94.0 | 2.0 | 9.0 | 18.0 | 41.0 | 6.0 | 17.0 | 17.0 | 95.0 | 13.0 | 15.0 |
| 18 | 16.0 | 18.0 | 9.0 | 27.0 | 51.0 | 88.0 | 7.0 | 150.0 | 3.0 | 10.0 | 7.0 | 8.0 | 4.0 | 11.0 | 16.0 | 17.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.477 | 0.262 | 0.176 | -0.739 | -0.18 | 0.341 | -1.65 | -0.667 | 0.289 | 0.907 | 0.526 | -0.023 | -1.855 | 0.262 | -0.224 | -0.318 |
| 02 | 0.234 | -0.023 | -0.816 | -1.334 | 1.016 | 0.768 | -2.114 | 0.366 | 0.39 | 0.262 | -0.27 | -1.094 | -0.368 | -0.27 | -0.27 | -0.739 |
| 03 | 0.114 | 0.817 | 0.832 | -0.667 | 0.664 | 0.341 | -1.094 | 0.414 | -0.27 | -0.536 | -1.094 | -0.9 | -0.9 | -0.06 | -0.139 | -1.206 |
| 04 | 0.315 | -2.114 | 0.977 | -2.465 | 1.256 | -3.01 | -0.27 | -0.139 | 0.366 | -2.465 | 0.717 | -1.334 | -0.599 | -3.01 | 0.7 | -2.114 |
| 05 | -1.094 | -3.01 | 1.846 | -1.855 | -2.465 | -4.298 | -2.114 | -2.465 | -0.739 | -3.01 | 1.895 | -1.206 | -4.298 | -4.298 | 0.234 | -3.01 |
| 06 | -0.992 | -2.114 | -1.206 | -2.114 | -3.01 | -4.298 | -4.298 | -3.01 | 1.992 | 0.483 | 1.296 | 0.567 | -2.114 | -1.855 | -3.01 | -1.334 |
| 07 | 1.973 | -4.298 | -0.536 | -2.465 | 0.587 | -2.465 | -4.298 | -4.298 | 1.325 | -2.465 | -2.114 | -3.01 | 0.664 | -4.298 | -2.114 | -2.114 |
| 08 | -4.298 | 2.627 | -0.739 | -0.992 | -4.298 | -1.855 | -4.298 | -4.298 | -3.01 | -0.27 | -4.298 | -4.298 | -4.298 | -1.479 | -4.298 | -4.298 |
| 09 | -4.298 | -3.01 | -4.298 | -4.298 | 2.56 | -1.479 | -0.368 | 0.114 | -1.206 | -4.298 | -2.465 | -2.114 | -2.114 | -3.01 | -1.479 | -4.298 |
| 10 | 0.848 | 1.003 | 1.713 | 1.016 | -3.01 | -3.01 | -3.01 | -1.65 | -4.298 | -1.479 | -0.667 | -1.334 | -3.01 | -1.094 | -0.667 | -0.992 |
| 11 | 0.013 | -1.206 | -0.098 | -1.479 | 0.145 | -0.18 | -0.739 | -0.139 | 0.735 | 0.526 | 0.626 | -0.023 | -0.421 | 0.145 | -0.139 | -0.098 |
| 12 | 0.801 | 0.289 | 0.013 | -1.094 | 0.483 | 0.39 | -0.739 | -0.9 | 0.315 | 0.176 | 0.205 | -1.094 | -1.334 | 0.483 | -0.318 | -0.992 |
| 13 | -0.27 | 0.922 | -0.992 | 0.607 | 0.526 | 0.048 | -3.01 | 1.042 | 0.114 | -0.368 | -0.816 | 0.176 | -1.479 | -1.094 | -3.01 | -0.18 |
| 14 | -1.65 | -1.855 | 1.225 | -3.01 | 0.526 | -0.992 | 0.893 | -2.465 | -2.114 | -2.465 | -0.421 | -3.01 | -1.334 | -1.334 | 1.811 | -3.01 |
| 15 | -1.206 | -1.65 | -0.536 | 0.176 | -2.465 | -2.465 | -2.465 | -0.477 | 0.341 | 0.341 | 0.39 | 2.12 | -3.01 | -3.01 | -2.114 | -4.298 |
| 16 | -0.9 | -1.206 | -0.477 | -0.667 | -0.667 | -1.479 | -2.465 | -0.098 | -0.139 | -0.599 | -1.334 | -0.536 | 0.626 | 1.172 | 0.7 | 1.115 |
| 17 | -0.27 | 0.863 | -1.334 | -1.334 | -0.667 | 1.215 | -2.465 | -1.094 | -0.421 | 0.39 | -1.479 | -0.477 | -0.477 | 1.225 | -0.739 | -0.599 |
| 18 | -0.536 | -0.421 | -1.094 | -0.023 | 0.607 | 1.149 | -1.334 | 1.681 | -2.114 | -0.992 | -1.334 | -1.206 | -1.855 | -0.9 | -0.536 | -0.477 |