Model info
| Transcription factor | Isl1 | ||||||||
| Model | ISL1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 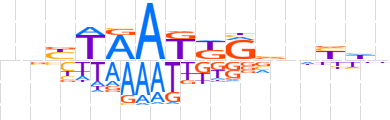 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 14 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nnYTAAKKRvhYhn | ||||||||
| Best auROC (human) | 0.838 | ||||||||
| Best auROC (mouse) | 0.918 | ||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | 13 | ||||||||
| Aligned words | 472 | ||||||||
| TF family | HD-LIM factors {3.1.5} | ||||||||
| TF subfamily | ISL {3.1.5.1} | ||||||||
| MGI | MGI:101791 | ||||||||
| EntrezGene | GeneID:16392 (SSTAR profile) | ||||||||
| UniProt ID | ISL1_MOUSE | ||||||||
| UniProt AC | P61372 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Isl1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 9.0 | 28.0 | 61.0 | 19.0 | 44.0 | 43.0 | 6.0 | 34.0 | 16.0 | 18.0 | 46.0 | 13.0 | 22.0 | 30.0 | 65.0 | 18.0 |
| 02 | 2.0 | 49.0 | 23.0 | 17.0 | 5.0 | 70.0 | 4.0 | 40.0 | 17.0 | 104.0 | 18.0 | 39.0 | 1.0 | 43.0 | 17.0 | 23.0 |
| 03 | 4.0 | 0.0 | 1.0 | 20.0 | 75.0 | 2.0 | 2.0 | 187.0 | 12.0 | 0.0 | 2.0 | 48.0 | 19.0 | 0.0 | 2.0 | 98.0 |
| 04 | 75.0 | 3.0 | 31.0 | 1.0 | 0.0 | 2.0 | 0.0 | 0.0 | 5.0 | 1.0 | 1.0 | 0.0 | 280.0 | 11.0 | 60.0 | 2.0 |
| 05 | 355.0 | 1.0 | 1.0 | 3.0 | 17.0 | 0.0 | 0.0 | 0.0 | 90.0 | 0.0 | 0.0 | 2.0 | 3.0 | 0.0 | 0.0 | 0.0 |
| 06 | 17.0 | 4.0 | 110.0 | 334.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 2.0 | 2.0 |
| 07 | 8.0 | 0.0 | 7.0 | 3.0 | 1.0 | 0.0 | 2.0 | 1.0 | 13.0 | 0.0 | 18.0 | 81.0 | 9.0 | 6.0 | 198.0 | 125.0 |
| 08 | 1.0 | 0.0 | 30.0 | 0.0 | 1.0 | 0.0 | 1.0 | 4.0 | 27.0 | 23.0 | 166.0 | 9.0 | 26.0 | 9.0 | 158.0 | 17.0 |
| 09 | 13.0 | 14.0 | 16.0 | 12.0 | 18.0 | 9.0 | 3.0 | 2.0 | 99.0 | 112.0 | 121.0 | 23.0 | 10.0 | 0.0 | 15.0 | 5.0 |
| 10 | 46.0 | 27.0 | 30.0 | 37.0 | 46.0 | 32.0 | 3.0 | 54.0 | 69.0 | 35.0 | 25.0 | 26.0 | 10.0 | 9.0 | 6.0 | 17.0 |
| 11 | 21.0 | 25.0 | 16.0 | 109.0 | 14.0 | 37.0 | 6.0 | 46.0 | 2.0 | 12.0 | 1.0 | 49.0 | 14.0 | 26.0 | 18.0 | 76.0 |
| 12 | 14.0 | 5.0 | 17.0 | 15.0 | 27.0 | 24.0 | 1.0 | 48.0 | 9.0 | 11.0 | 10.0 | 11.0 | 46.0 | 41.0 | 26.0 | 167.0 |
| 13 | 25.0 | 15.0 | 31.0 | 25.0 | 25.0 | 15.0 | 3.0 | 38.0 | 23.0 | 13.0 | 7.0 | 11.0 | 65.0 | 42.0 | 39.0 | 95.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.158 | -0.051 | 0.72 | -0.433 | 0.396 | 0.373 | -1.543 | 0.14 | -0.601 | -0.486 | 0.44 | -0.803 | -0.289 | 0.017 | 0.783 | -0.486 |
| 02 | -2.528 | 0.502 | -0.245 | -0.542 | -1.714 | 0.857 | -1.919 | 0.301 | -0.542 | 1.251 | -0.486 | 0.276 | -3.072 | 0.373 | -0.542 | -0.245 |
| 03 | -1.919 | -4.352 | -3.072 | -0.383 | 0.925 | -2.528 | -2.528 | 1.836 | -0.881 | -4.352 | -2.528 | 0.482 | -0.433 | -4.352 | -2.528 | 1.192 |
| 04 | 0.925 | -2.178 | 0.049 | -3.072 | -4.352 | -2.528 | -4.352 | -4.352 | -1.714 | -3.072 | -3.072 | -4.352 | 2.239 | -0.965 | 0.703 | -2.528 |
| 05 | 2.476 | -3.072 | -3.072 | -2.178 | -0.542 | -4.352 | -4.352 | -4.352 | 1.107 | -4.352 | -4.352 | -2.528 | -2.178 | -4.352 | -4.352 | -4.352 |
| 06 | -0.542 | -1.919 | 1.307 | 2.415 | -4.352 | -4.352 | -4.352 | -3.072 | -4.352 | -4.352 | -4.352 | -3.072 | -3.072 | -4.352 | -2.528 | -2.528 |
| 07 | -1.271 | -4.352 | -1.398 | -2.178 | -3.072 | -4.352 | -2.528 | -3.072 | -0.803 | -4.352 | -0.486 | 1.002 | -1.158 | -1.543 | 1.893 | 1.434 |
| 08 | -3.072 | -4.352 | 0.017 | -4.352 | -3.072 | -4.352 | -3.072 | -1.919 | -0.087 | -0.245 | 1.717 | -1.158 | -0.125 | -1.158 | 1.668 | -0.542 |
| 09 | -0.803 | -0.731 | -0.601 | -0.881 | -0.486 | -1.158 | -2.178 | -2.528 | 1.202 | 1.325 | 1.402 | -0.245 | -1.057 | -4.352 | -0.664 | -1.714 |
| 10 | 0.44 | -0.087 | 0.017 | 0.224 | 0.44 | 0.08 | -2.178 | 0.599 | 0.842 | 0.169 | -0.163 | -0.125 | -1.057 | -1.158 | -1.543 | -0.542 |
| 11 | -0.335 | -0.163 | -0.601 | 1.298 | -0.731 | 0.224 | -1.543 | 0.44 | -2.528 | -0.881 | -3.072 | 0.502 | -0.731 | -0.125 | -0.486 | 0.938 |
| 12 | -0.731 | -1.714 | -0.542 | -0.664 | -0.087 | -0.203 | -3.072 | 0.482 | -1.158 | -0.965 | -1.057 | -0.965 | 0.44 | 0.326 | -0.125 | 1.723 |
| 13 | -0.163 | -0.664 | 0.049 | -0.163 | -0.163 | -0.664 | -2.178 | 0.25 | -0.245 | -0.803 | -1.398 | -0.965 | 0.783 | 0.349 | 0.276 | 1.161 |