Model info
| Transcription factor | Lhx6 | ||||||||
| Model | LHX6_MOUSE.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 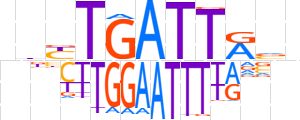 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 11 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbYTGATTRvn | ||||||||
| Best auROC (human) | 0.337 | ||||||||
| Best auROC (mouse) | 0.932 | ||||||||
| Peak sets in benchmark (human) | 1 | ||||||||
| Peak sets in benchmark (mouse) | 2 | ||||||||
| Aligned words | 445 | ||||||||
| TF family | HD-LIM factors {3.1.5} | ||||||||
| TF subfamily | Lhx-6-like factors {3.1.5.5} | ||||||||
| MGI | MGI:1306803 | ||||||||
| EntrezGene | GeneID:16874 (SSTAR profile) | ||||||||
| UniProt ID | LHX6_MOUSE | ||||||||
| UniProt AC | Q9R1R0 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Lhx6 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 10.0 | 25.0 | 36.0 | 32.0 | 21.0 | 16.0 | 12.0 | 59.0 | 12.0 | 31.0 | 21.0 | 39.0 | 8.0 | 28.0 | 59.0 | 35.0 |
| 02 | 1.0 | 33.0 | 13.0 | 4.0 | 5.0 | 47.0 | 6.0 | 42.0 | 7.0 | 93.0 | 7.0 | 21.0 | 7.0 | 79.0 | 24.0 | 55.0 |
| 03 | 0.0 | 0.0 | 0.0 | 20.0 | 2.0 | 0.0 | 0.0 | 250.0 | 0.0 | 0.0 | 0.0 | 50.0 | 0.0 | 0.0 | 1.0 | 121.0 |
| 04 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 48.0 | 1.0 | 392.0 | 0.0 |
| 05 | 48.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 393.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 4.0 | 1.0 | 437.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 17.0 | 421.0 |
| 08 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 16.0 | 0.0 | 1.0 | 0.0 | 242.0 | 0.0 | 175.0 | 9.0 |
| 09 | 61.0 | 77.0 | 103.0 | 17.0 | 0.0 | 0.0 | 0.0 | 0.0 | 23.0 | 100.0 | 39.0 | 15.0 | 3.0 | 4.0 | 2.0 | 0.0 |
| 10 | 15.0 | 28.0 | 23.0 | 21.0 | 78.0 | 31.0 | 15.0 | 57.0 | 32.0 | 54.0 | 33.0 | 25.0 | 8.0 | 2.0 | 8.0 | 14.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.997 | -0.103 | 0.257 | 0.141 | -0.274 | -0.541 | -0.821 | 0.747 | -0.821 | 0.109 | -0.274 | 0.336 | -1.211 | 0.009 | 0.747 | 0.229 |
| 02 | -3.014 | 0.171 | -0.743 | -1.86 | -1.654 | 0.521 | -1.484 | 0.41 | -1.338 | 1.2 | -1.338 | -0.274 | -1.338 | 1.037 | -0.143 | 0.677 |
| 03 | -4.302 | -4.302 | -4.302 | -0.322 | -2.469 | -4.302 | -4.302 | 2.186 | -4.302 | -4.302 | -4.302 | 0.583 | -4.302 | -4.302 | -3.014 | 1.462 |
| 04 | -4.302 | -4.302 | -2.469 | -4.302 | -4.302 | -4.302 | -4.302 | -4.302 | -4.302 | -4.302 | -3.014 | -4.302 | 0.542 | -3.014 | 2.635 | -4.302 |
| 05 | 0.542 | -4.302 | -4.302 | -4.302 | -3.014 | -4.302 | -4.302 | -4.302 | 2.638 | -4.302 | -2.469 | -4.302 | -4.302 | -4.302 | -4.302 | -4.302 |
| 06 | -4.302 | -1.86 | -3.014 | 2.744 | -4.302 | -4.302 | -4.302 | -4.302 | -4.302 | -4.302 | -4.302 | -2.469 | -4.302 | -4.302 | -4.302 | -4.302 |
| 07 | -4.302 | -4.302 | -4.302 | -4.302 | -4.302 | -4.302 | -4.302 | -1.86 | -4.302 | -4.302 | -4.302 | -3.014 | -3.014 | -4.302 | -0.481 | 2.707 |
| 08 | -4.302 | -4.302 | -3.014 | -4.302 | -4.302 | -4.302 | -4.302 | -4.302 | -0.541 | -4.302 | -3.014 | -4.302 | 2.154 | -4.302 | 1.83 | -1.098 |
| 09 | 0.78 | 1.012 | 1.302 | -0.481 | -4.302 | -4.302 | -4.302 | -4.302 | -0.185 | 1.272 | 0.336 | -0.604 | -2.119 | -1.86 | -2.469 | -4.302 |
| 10 | -0.604 | 0.009 | -0.185 | -0.274 | 1.025 | 0.109 | -0.604 | 0.713 | 0.141 | 0.659 | 0.171 | -0.103 | -1.211 | -2.469 | -1.211 | -0.671 |