Model info
| Transcription factor | MAFF (GeneCards) | ||||||||
| Model | MAFF_HUMAN.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 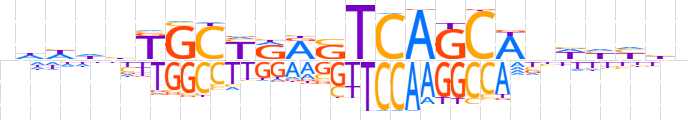 | ||||||||
| LOGO (reverse complement) | 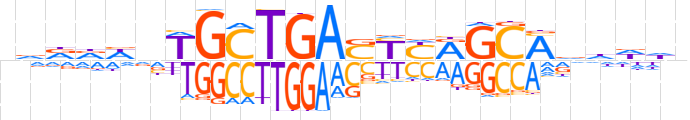 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 24 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nhhhbKGCYRRSTCAGCWndhhhn | ||||||||
| Best auROC (human) | 0.489 | ||||||||
| Best auROC (mouse) | 0.982 | ||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | 15 | ||||||||
| Aligned words | 433 | ||||||||
| TF family | Maf-related factors {1.1.3} | ||||||||
| TF subfamily | Small Maf factors {1.1.3.2} | ||||||||
| HGNC | HGNC:6780 | ||||||||
| EntrezGene | GeneID:23764 (SSTAR profile) | ||||||||
| UniProt ID | MAFF_HUMAN | ||||||||
| UniProt AC | Q9ULX9 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | MAFF expression | ||||||||
| ReMap ChIP-seq dataset list | MAFF datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 79.0 | 12.0 | 14.0 | 47.0 | 38.0 | 12.0 | 3.0 | 24.0 | 46.0 | 15.0 | 14.0 | 19.0 | 38.0 | 12.0 | 15.0 | 45.0 |
| 02 | 108.0 | 13.0 | 16.0 | 64.0 | 19.0 | 11.0 | 1.0 | 20.0 | 19.0 | 7.0 | 8.0 | 12.0 | 42.0 | 10.0 | 5.0 | 78.0 |
| 03 | 84.0 | 17.0 | 27.0 | 60.0 | 20.0 | 2.0 | 2.0 | 17.0 | 9.0 | 7.0 | 8.0 | 6.0 | 27.0 | 28.0 | 16.0 | 103.0 |
| 04 | 27.0 | 41.0 | 35.0 | 37.0 | 8.0 | 16.0 | 0.0 | 30.0 | 9.0 | 13.0 | 13.0 | 18.0 | 13.0 | 58.0 | 33.0 | 82.0 |
| 05 | 4.0 | 3.0 | 5.0 | 45.0 | 3.0 | 6.0 | 2.0 | 117.0 | 7.0 | 6.0 | 7.0 | 61.0 | 4.0 | 9.0 | 32.0 | 122.0 |
| 06 | 2.0 | 1.0 | 14.0 | 1.0 | 4.0 | 4.0 | 15.0 | 1.0 | 4.0 | 26.0 | 13.0 | 3.0 | 2.0 | 10.0 | 331.0 | 2.0 |
| 07 | 1.0 | 8.0 | 2.0 | 1.0 | 5.0 | 24.0 | 2.0 | 10.0 | 9.0 | 341.0 | 4.0 | 19.0 | 0.0 | 3.0 | 1.0 | 3.0 |
| 08 | 1.0 | 3.0 | 2.0 | 9.0 | 37.0 | 40.0 | 5.0 | 294.0 | 0.0 | 2.0 | 2.0 | 5.0 | 2.0 | 12.0 | 11.0 | 8.0 |
| 09 | 11.0 | 10.0 | 14.0 | 5.0 | 21.0 | 8.0 | 13.0 | 15.0 | 9.0 | 1.0 | 5.0 | 5.0 | 14.0 | 4.0 | 271.0 | 27.0 |
| 10 | 17.0 | 4.0 | 29.0 | 5.0 | 17.0 | 0.0 | 3.0 | 3.0 | 257.0 | 14.0 | 11.0 | 21.0 | 32.0 | 0.0 | 15.0 | 5.0 |
| 11 | 6.0 | 86.0 | 216.0 | 15.0 | 6.0 | 8.0 | 4.0 | 0.0 | 1.0 | 31.0 | 25.0 | 1.0 | 1.0 | 3.0 | 29.0 | 1.0 |
| 12 | 0.0 | 1.0 | 2.0 | 11.0 | 0.0 | 0.0 | 0.0 | 128.0 | 0.0 | 1.0 | 3.0 | 270.0 | 0.0 | 0.0 | 2.0 | 15.0 |
| 13 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 2.0 | 12.0 | 401.0 | 5.0 | 6.0 |
| 14 | 9.0 | 0.0 | 3.0 | 0.0 | 403.0 | 0.0 | 2.0 | 3.0 | 2.0 | 0.0 | 2.0 | 1.0 | 0.0 | 1.0 | 4.0 | 3.0 |
| 15 | 2.0 | 4.0 | 340.0 | 68.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 8.0 | 2.0 | 0.0 | 0.0 | 6.0 | 1.0 |
| 16 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 1.0 | 0.0 | 350.0 | 2.0 | 2.0 | 0.0 | 55.0 | 9.0 | 8.0 |
| 17 | 0.0 | 0.0 | 0.0 | 0.0 | 337.0 | 8.0 | 22.0 | 44.0 | 2.0 | 1.0 | 5.0 | 3.0 | 1.0 | 1.0 | 2.0 | 7.0 |
| 18 | 57.0 | 89.0 | 69.0 | 125.0 | 3.0 | 1.0 | 0.0 | 6.0 | 5.0 | 7.0 | 6.0 | 11.0 | 7.0 | 15.0 | 16.0 | 16.0 |
| 19 | 37.0 | 0.0 | 6.0 | 29.0 | 23.0 | 10.0 | 4.0 | 75.0 | 27.0 | 6.0 | 17.0 | 41.0 | 16.0 | 22.0 | 18.0 | 102.0 |
| 20 | 43.0 | 11.0 | 16.0 | 33.0 | 4.0 | 21.0 | 0.0 | 13.0 | 6.0 | 8.0 | 8.0 | 23.0 | 21.0 | 45.0 | 10.0 | 171.0 |
| 21 | 16.0 | 19.0 | 5.0 | 34.0 | 17.0 | 36.0 | 1.0 | 31.0 | 8.0 | 10.0 | 3.0 | 13.0 | 15.0 | 63.0 | 13.0 | 149.0 |
| 22 | 12.0 | 11.0 | 13.0 | 20.0 | 33.0 | 33.0 | 1.0 | 61.0 | 1.0 | 8.0 | 6.0 | 7.0 | 31.0 | 51.0 | 32.0 | 113.0 |
| 23 | 11.0 | 15.0 | 35.0 | 16.0 | 26.0 | 33.0 | 6.0 | 38.0 | 9.0 | 8.0 | 23.0 | 12.0 | 26.0 | 52.0 | 62.0 | 61.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 1.062 | -0.796 | -0.646 | 0.546 | 0.335 | -0.796 | -2.094 | -0.118 | 0.525 | -0.579 | -0.646 | -0.348 | 0.335 | -0.796 | -0.579 | 0.503 |
| 02 | 1.374 | -0.718 | -0.516 | 0.853 | -0.348 | -0.88 | -2.99 | -0.298 | -0.348 | -1.313 | -1.186 | -0.796 | 0.435 | -0.972 | -1.629 | 1.049 |
| 03 | 1.123 | -0.457 | -0.002 | 0.789 | -0.298 | -2.445 | -2.445 | -0.457 | -1.074 | -1.313 | -1.186 | -1.459 | -0.002 | 0.034 | -0.516 | 1.326 |
| 04 | -0.002 | 0.411 | 0.254 | 0.309 | -1.186 | -0.516 | -4.281 | 0.102 | -1.074 | -0.718 | -0.718 | -0.401 | -0.718 | 0.755 | 0.196 | 1.099 |
| 05 | -1.835 | -2.094 | -1.629 | 0.503 | -2.094 | -1.459 | -2.445 | 1.453 | -1.313 | -1.459 | -1.313 | 0.805 | -1.835 | -1.074 | 0.165 | 1.495 |
| 06 | -2.445 | -2.99 | -0.646 | -2.99 | -1.835 | -1.835 | -0.579 | -2.99 | -1.835 | -0.039 | -0.718 | -2.094 | -2.445 | -0.972 | 2.491 | -2.445 |
| 07 | -2.99 | -1.186 | -2.445 | -2.99 | -1.629 | -0.118 | -2.445 | -0.972 | -1.074 | 2.521 | -1.835 | -0.348 | -4.281 | -2.094 | -2.99 | -2.094 |
| 08 | -2.99 | -2.094 | -2.445 | -1.074 | 0.309 | 0.386 | -1.629 | 2.373 | -4.281 | -2.445 | -2.445 | -1.629 | -2.445 | -0.796 | -0.88 | -1.186 |
| 09 | -0.88 | -0.972 | -0.646 | -1.629 | -0.25 | -1.186 | -0.718 | -0.579 | -1.074 | -2.99 | -1.629 | -1.629 | -0.646 | -1.835 | 2.291 | -0.002 |
| 10 | -0.457 | -1.835 | 0.068 | -1.629 | -0.457 | -4.281 | -2.094 | -2.094 | 2.238 | -0.646 | -0.88 | -0.25 | 0.165 | -4.281 | -0.579 | -1.629 |
| 11 | -1.459 | 1.147 | 2.065 | -0.579 | -1.459 | -1.186 | -1.835 | -4.281 | -2.99 | 0.134 | -0.078 | -2.99 | -2.99 | -2.094 | 0.068 | -2.99 |
| 12 | -4.281 | -2.99 | -2.445 | -0.88 | -4.281 | -4.281 | -4.281 | 1.543 | -4.281 | -2.99 | -2.094 | 2.288 | -4.281 | -4.281 | -2.445 | -0.579 |
| 13 | -4.281 | -4.281 | -4.281 | -4.281 | -4.281 | -2.445 | -4.281 | -4.281 | -4.281 | -1.629 | -4.281 | -2.445 | -0.796 | 2.683 | -1.629 | -1.459 |
| 14 | -1.074 | -4.281 | -2.094 | -4.281 | 2.688 | -4.281 | -2.445 | -2.094 | -2.445 | -4.281 | -2.445 | -2.99 | -4.281 | -2.99 | -1.835 | -2.094 |
| 15 | -2.445 | -1.835 | 2.518 | 0.913 | -4.281 | -4.281 | -4.281 | -2.99 | -4.281 | -2.99 | -1.186 | -2.445 | -4.281 | -4.281 | -1.459 | -2.99 |
| 16 | -4.281 | -2.445 | -4.281 | -4.281 | -4.281 | -1.835 | -4.281 | -2.99 | -4.281 | 2.547 | -2.445 | -2.445 | -4.281 | 0.702 | -1.074 | -1.186 |
| 17 | -4.281 | -4.281 | -4.281 | -4.281 | 2.509 | -1.186 | -0.204 | 0.481 | -2.445 | -2.99 | -1.629 | -2.094 | -2.99 | -2.99 | -2.445 | -1.313 |
| 18 | 0.738 | 1.181 | 0.928 | 1.519 | -2.094 | -2.99 | -4.281 | -1.459 | -1.629 | -1.313 | -1.459 | -0.88 | -1.313 | -0.579 | -0.516 | -0.516 |
| 19 | 0.309 | -4.281 | -1.459 | 0.068 | -0.16 | -0.972 | -1.835 | 1.01 | -0.002 | -1.459 | -0.457 | 0.411 | -0.516 | -0.204 | -0.401 | 1.317 |
| 20 | 0.458 | -0.88 | -0.516 | 0.196 | -1.835 | -0.25 | -4.281 | -0.718 | -1.459 | -1.186 | -1.186 | -0.16 | -0.25 | 0.503 | -0.972 | 1.832 |
| 21 | -0.516 | -0.348 | -1.629 | 0.225 | -0.457 | 0.282 | -2.99 | 0.134 | -1.186 | -0.972 | -2.094 | -0.718 | -0.579 | 0.837 | -0.718 | 1.694 |
| 22 | -0.796 | -0.88 | -0.718 | -0.298 | 0.196 | 0.196 | -2.99 | 0.805 | -2.99 | -1.186 | -1.459 | -1.313 | 0.134 | 0.627 | 0.165 | 1.419 |
| 23 | -0.88 | -0.579 | 0.254 | -0.516 | -0.039 | 0.196 | -1.459 | 0.335 | -1.074 | -1.186 | -0.16 | -0.796 | -0.039 | 0.646 | 0.821 | 0.805 |