Model info
| Transcription factor | Myc | ||||||||
| Model | MYC_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 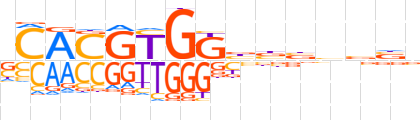 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bvvvvvvMCACGTGb | ||||||||
| Best auROC (human) | 0.909 | ||||||||
| Best auROC (mouse) | 0.963 | ||||||||
| Peak sets in benchmark (human) | 265 | ||||||||
| Peak sets in benchmark (mouse) | 126 | ||||||||
| Aligned words | 450 | ||||||||
| TF family | bHLH-ZIP factors {1.2.6} | ||||||||
| TF subfamily | Myc / Max factors {1.2.6.5} | ||||||||
| MGI | MGI:97250 | ||||||||
| EntrezGene | GeneID:17869 (SSTAR profile) | ||||||||
| UniProt ID | MYC_MOUSE | ||||||||
| UniProt AC | P01108 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Myc expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 16.0 | 20.0 | 22.0 | 5.0 | 24.0 | 91.0 | 28.0 | 18.0 | 23.0 | 88.0 | 28.0 | 8.0 | 5.0 | 51.0 | 7.0 | 16.0 |
| 02 | 18.0 | 31.0 | 15.0 | 4.0 | 44.0 | 96.0 | 63.0 | 47.0 | 16.0 | 44.0 | 20.0 | 5.0 | 6.0 | 19.0 | 20.0 | 2.0 |
| 03 | 19.0 | 17.0 | 35.0 | 13.0 | 38.0 | 48.0 | 72.0 | 32.0 | 24.0 | 42.0 | 39.0 | 13.0 | 5.0 | 25.0 | 22.0 | 6.0 |
| 04 | 14.0 | 33.0 | 28.0 | 11.0 | 22.0 | 54.0 | 33.0 | 23.0 | 17.0 | 101.0 | 40.0 | 10.0 | 6.0 | 36.0 | 16.0 | 6.0 |
| 05 | 17.0 | 7.0 | 32.0 | 3.0 | 57.0 | 80.0 | 79.0 | 8.0 | 32.0 | 39.0 | 45.0 | 1.0 | 12.0 | 11.0 | 25.0 | 2.0 |
| 06 | 35.0 | 25.0 | 55.0 | 3.0 | 37.0 | 22.0 | 59.0 | 19.0 | 51.0 | 43.0 | 83.0 | 4.0 | 3.0 | 7.0 | 3.0 | 1.0 |
| 07 | 25.0 | 83.0 | 18.0 | 0.0 | 19.0 | 64.0 | 13.0 | 1.0 | 22.0 | 176.0 | 1.0 | 1.0 | 3.0 | 21.0 | 2.0 | 1.0 |
| 08 | 3.0 | 66.0 | 0.0 | 0.0 | 7.0 | 332.0 | 2.0 | 3.0 | 2.0 | 32.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 |
| 09 | 7.0 | 0.0 | 5.0 | 0.0 | 341.0 | 3.0 | 63.0 | 26.0 | 2.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 |
| 10 | 1.0 | 272.0 | 5.0 | 75.0 | 0.0 | 1.0 | 0.0 | 2.0 | 1.0 | 65.0 | 1.0 | 1.0 | 0.0 | 22.0 | 2.0 | 2.0 |
| 11 | 0.0 | 1.0 | 1.0 | 0.0 | 15.0 | 29.0 | 304.0 | 12.0 | 0.0 | 3.0 | 5.0 | 0.0 | 6.0 | 13.0 | 55.0 | 6.0 |
| 12 | 1.0 | 2.0 | 1.0 | 17.0 | 1.0 | 3.0 | 1.0 | 41.0 | 10.0 | 64.0 | 2.0 | 289.0 | 0.0 | 3.0 | 1.0 | 14.0 |
| 13 | 0.0 | 2.0 | 7.0 | 3.0 | 0.0 | 4.0 | 64.0 | 4.0 | 0.0 | 1.0 | 3.0 | 1.0 | 3.0 | 15.0 | 306.0 | 37.0 |
| 14 | 1.0 | 0.0 | 2.0 | 0.0 | 6.0 | 11.0 | 0.0 | 5.0 | 43.0 | 163.0 | 134.0 | 40.0 | 6.0 | 6.0 | 21.0 | 12.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.554 | -0.335 | -0.242 | -1.667 | -0.156 | 1.165 | -0.004 | -0.439 | -0.198 | 1.132 | -0.004 | -1.224 | -1.667 | 0.589 | -1.351 | -0.554 |
| 02 | -0.439 | 0.096 | -0.617 | -1.873 | 0.443 | 1.218 | 0.799 | 0.508 | -0.554 | 0.443 | -0.335 | -1.667 | -1.497 | -0.386 | -0.335 | -2.482 |
| 03 | -0.386 | -0.495 | 0.216 | -0.756 | 0.297 | 0.529 | 0.932 | 0.127 | -0.156 | 0.397 | 0.323 | -0.756 | -1.667 | -0.116 | -0.242 | -1.497 |
| 04 | -0.684 | 0.158 | -0.004 | -0.918 | -0.242 | 0.646 | 0.158 | -0.198 | -0.495 | 1.269 | 0.348 | -1.01 | -1.497 | 0.244 | -0.554 | -1.497 |
| 05 | -0.495 | -1.351 | 0.127 | -2.132 | 0.7 | 1.037 | 1.024 | -1.224 | 0.127 | 0.323 | 0.465 | -3.027 | -0.834 | -0.918 | -0.116 | -2.482 |
| 06 | 0.216 | -0.116 | 0.664 | -2.132 | 0.271 | -0.242 | 0.734 | -0.386 | 0.589 | 0.42 | 1.073 | -1.873 | -2.132 | -1.351 | -2.132 | -3.027 |
| 07 | -0.116 | 1.073 | -0.439 | -4.313 | -0.386 | 0.815 | -0.756 | -3.027 | -0.242 | 1.823 | -3.027 | -3.027 | -2.132 | -0.288 | -2.482 | -3.027 |
| 08 | -2.132 | 0.845 | -4.313 | -4.313 | -1.351 | 2.456 | -2.482 | -2.132 | -2.482 | 0.127 | -4.313 | -4.313 | -4.313 | -2.132 | -4.313 | -4.313 |
| 09 | -1.351 | -4.313 | -1.667 | -4.313 | 2.483 | -2.132 | 0.799 | -0.077 | -2.482 | -4.313 | -4.313 | -4.313 | -2.132 | -4.313 | -4.313 | -4.313 |
| 10 | -3.027 | 2.257 | -1.667 | 0.972 | -4.313 | -3.027 | -4.313 | -2.482 | -3.027 | 0.83 | -3.027 | -3.027 | -4.313 | -0.242 | -2.482 | -2.482 |
| 11 | -4.313 | -3.027 | -3.027 | -4.313 | -0.617 | 0.03 | 2.368 | -0.834 | -4.313 | -2.132 | -1.667 | -4.313 | -1.497 | -0.756 | 0.664 | -1.497 |
| 12 | -3.027 | -2.482 | -3.027 | -0.495 | -3.027 | -2.132 | -3.027 | 0.373 | -1.01 | 0.815 | -2.482 | 2.318 | -4.313 | -2.132 | -3.027 | -0.684 |
| 13 | -4.313 | -2.482 | -1.351 | -2.132 | -4.313 | -1.873 | 0.815 | -1.873 | -4.313 | -3.027 | -2.132 | -3.027 | -2.132 | -0.617 | 2.375 | 0.271 |
| 14 | -3.027 | -4.313 | -2.482 | -4.313 | -1.497 | -0.918 | -4.313 | -1.667 | 0.42 | 1.746 | 1.551 | 0.348 | -1.497 | -1.497 | -0.288 | -0.834 |