Model info
| Transcription factor | Nkx2-1 | ||||||||
| Model | NKX21_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 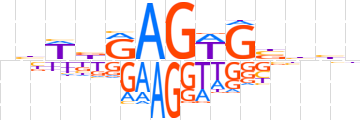 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 13 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbWbRAGWGbbhn | ||||||||
| Best auROC (human) | 0.905 | ||||||||
| Best auROC (mouse) | 0.937 | ||||||||
| Peak sets in benchmark (human) | 14 | ||||||||
| Peak sets in benchmark (mouse) | 9 | ||||||||
| Aligned words | 502 | ||||||||
| TF family | NK-related factors {3.1.2} | ||||||||
| TF subfamily | NK-2.1 {3.1.2.14} | ||||||||
| MGI | MGI:108067 | ||||||||
| EntrezGene | GeneID:21869 (SSTAR profile) | ||||||||
| UniProt ID | NKX21_MOUSE | ||||||||
| UniProt AC | P50220 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Nkx2-1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 14.0 | 38.0 | 27.0 | 21.0 | 27.0 | 51.0 | 16.0 | 33.0 | 5.0 | 43.0 | 31.0 | 31.0 | 5.0 | 59.0 | 36.0 | 43.0 |
| 02 | 6.0 | 12.0 | 12.0 | 21.0 | 32.0 | 14.0 | 2.0 | 143.0 | 14.0 | 17.0 | 2.0 | 77.0 | 12.0 | 10.0 | 6.0 | 100.0 |
| 03 | 7.0 | 6.0 | 13.0 | 38.0 | 6.0 | 2.0 | 2.0 | 43.0 | 2.0 | 0.0 | 2.0 | 18.0 | 33.0 | 54.0 | 114.0 | 140.0 |
| 04 | 8.0 | 6.0 | 34.0 | 0.0 | 31.0 | 12.0 | 19.0 | 0.0 | 36.0 | 6.0 | 88.0 | 1.0 | 23.0 | 8.0 | 208.0 | 0.0 |
| 05 | 98.0 | 0.0 | 0.0 | 0.0 | 32.0 | 0.0 | 0.0 | 0.0 | 343.0 | 0.0 | 5.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 06 | 1.0 | 2.0 | 469.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 07 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 139.0 | 1.0 | 23.0 | 312.0 | 1.0 | 0.0 | 0.0 | 1.0 |
| 08 | 3.0 | 1.0 | 134.0 | 4.0 | 0.0 | 0.0 | 0.0 | 1.0 | 5.0 | 0.0 | 18.0 | 0.0 | 37.0 | 2.0 | 265.0 | 10.0 |
| 09 | 1.0 | 30.0 | 7.0 | 7.0 | 1.0 | 0.0 | 0.0 | 2.0 | 9.0 | 138.0 | 169.0 | 101.0 | 0.0 | 8.0 | 2.0 | 5.0 |
| 10 | 0.0 | 3.0 | 5.0 | 3.0 | 31.0 | 66.0 | 4.0 | 75.0 | 27.0 | 65.0 | 44.0 | 42.0 | 15.0 | 41.0 | 22.0 | 37.0 |
| 11 | 16.0 | 26.0 | 14.0 | 17.0 | 32.0 | 63.0 | 6.0 | 74.0 | 9.0 | 32.0 | 12.0 | 22.0 | 21.0 | 39.0 | 20.0 | 77.0 |
| 12 | 23.0 | 31.0 | 16.0 | 8.0 | 28.0 | 25.0 | 7.0 | 100.0 | 14.0 | 13.0 | 18.0 | 7.0 | 29.0 | 60.0 | 53.0 | 48.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.748 | 0.234 | -0.104 | -0.351 | -0.104 | 0.525 | -0.618 | 0.094 | -1.73 | 0.356 | 0.032 | 0.032 | -1.73 | 0.67 | 0.18 | 0.356 |
| 02 | -1.56 | -0.897 | -0.897 | -0.351 | 0.064 | -0.748 | -2.544 | 1.552 | -0.748 | -0.558 | -2.544 | 0.935 | -0.897 | -1.074 | -1.56 | 1.195 |
| 03 | -1.414 | -1.56 | -0.82 | 0.234 | -1.56 | -2.544 | -2.544 | 0.356 | -2.544 | -4.366 | -2.544 | -0.502 | 0.094 | 0.582 | 1.326 | 1.53 |
| 04 | -1.287 | -1.56 | 0.124 | -4.366 | 0.032 | -0.897 | -0.449 | -4.366 | 0.18 | -1.56 | 1.068 | -3.088 | -0.262 | -1.287 | 1.925 | -4.366 |
| 05 | 1.175 | -4.366 | -4.366 | -4.366 | 0.064 | -4.366 | -4.366 | -4.366 | 2.425 | -4.366 | -1.73 | -3.088 | -3.088 | -4.366 | -4.366 | -4.366 |
| 06 | -3.088 | -2.544 | 2.737 | -2.544 | -4.366 | -4.366 | -4.366 | -4.366 | -4.366 | -4.366 | -1.73 | -4.366 | -4.366 | -4.366 | -3.088 | -4.366 |
| 07 | -3.088 | -4.366 | -4.366 | -4.366 | -3.088 | -4.366 | -4.366 | -3.088 | 1.523 | -3.088 | -0.262 | 2.33 | -3.088 | -4.366 | -4.366 | -3.088 |
| 08 | -2.194 | -3.088 | 1.487 | -1.936 | -4.366 | -4.366 | -4.366 | -3.088 | -1.73 | -4.366 | -0.502 | -4.366 | 0.207 | -2.544 | 2.167 | -1.074 |
| 09 | -3.088 | 0.0 | -1.414 | -1.414 | -3.088 | -4.366 | -4.366 | -2.544 | -1.175 | 1.516 | 1.718 | 1.205 | -4.366 | -1.287 | -2.544 | -1.73 |
| 10 | -4.366 | -2.194 | -1.73 | -2.194 | 0.032 | 0.782 | -1.936 | 0.909 | -0.104 | 0.766 | 0.379 | 0.333 | -0.681 | 0.309 | -0.306 | 0.207 |
| 11 | -0.618 | -0.141 | -0.748 | -0.558 | 0.064 | 0.735 | -1.56 | 0.895 | -1.175 | 0.064 | -0.897 | -0.306 | -0.351 | 0.259 | -0.399 | 0.935 |
| 12 | -0.262 | 0.032 | -0.618 | -1.287 | -0.068 | -0.18 | -1.414 | 1.195 | -0.748 | -0.82 | -0.502 | -1.414 | -0.033 | 0.687 | 0.564 | 0.465 |