Model info
| Transcription factor | NR5A2 (GeneCards) | ||||||||
| Model | NR5A2_HUMAN.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 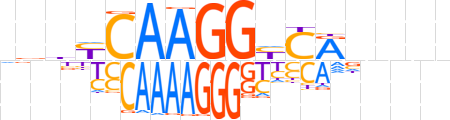 | ||||||||
| LOGO (reverse complement) | 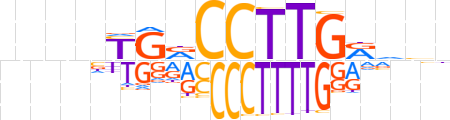 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 16 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndbYCAAGGYCRnnnn | ||||||||
| Best auROC (human) | 0.526 | ||||||||
| Best auROC (mouse) | 0.957 | ||||||||
| Peak sets in benchmark (human) | 4 | ||||||||
| Peak sets in benchmark (mouse) | 8 | ||||||||
| Aligned words | 501 | ||||||||
| TF family | FTZ-F1-related receptors (NR5) {2.1.5} | ||||||||
| TF subfamily | LRH-1 (NR5A2) {2.1.5.0.2} | ||||||||
| HGNC | HGNC:7984 | ||||||||
| EntrezGene | GeneID:2494 (SSTAR profile) | ||||||||
| UniProt ID | NR5A2_HUMAN | ||||||||
| UniProt AC | O00482 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | NR5A2 expression | ||||||||
| ReMap ChIP-seq dataset list | NR5A2 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 44.527 | 18.993 | 103.56 | 21.204 | 28.71 | 6.611 | 2.857 | 40.885 | 19.211 | 25.995 | 32.86 | 25.994 | 17.746 | 11.925 | 57.112 | 41.81 |
| 02 | 19.956 | 18.795 | 37.289 | 34.154 | 9.022 | 22.575 | 0.922 | 31.005 | 16.779 | 43.183 | 53.535 | 82.891 | 9.506 | 39.064 | 29.345 | 51.979 |
| 03 | 8.572 | 20.429 | 4.192 | 22.069 | 6.951 | 59.528 | 0.0 | 57.139 | 1.935 | 56.4 | 8.235 | 54.52 | 12.413 | 36.435 | 4.21 | 146.97 |
| 04 | 0.0 | 28.958 | 0.913 | 0.0 | 0.899 | 164.551 | 5.311 | 2.03 | 0.0 | 13.691 | 2.947 | 0.0 | 1.007 | 258.94 | 19.854 | 0.897 |
| 05 | 1.907 | 0.0 | 0.0 | 0.0 | 461.171 | 1.859 | 2.202 | 0.907 | 28.134 | 0.0 | 0.891 | 0.0 | 2.03 | 0.0 | 0.0 | 0.897 |
| 06 | 473.391 | 0.859 | 15.815 | 3.177 | 1.859 | 0.0 | 0.0 | 0.0 | 3.094 | 0.0 | 0.0 | 0.0 | 1.805 | 0.0 | 0.0 | 0.0 |
| 07 | 4.72 | 0.0 | 475.429 | 0.0 | 0.0 | 0.0 | 0.859 | 0.0 | 0.0 | 0.0 | 14.922 | 0.893 | 0.0 | 0.0 | 2.328 | 0.849 |
| 08 | 0.0 | 0.0 | 4.72 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.773 | 0.0 | 491.765 | 0.0 | 0.0 | 0.0 | 1.742 | 0.0 |
| 09 | 0.899 | 0.873 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 44.702 | 181.406 | 21.751 | 250.368 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 1.782 | 40.076 | 1.762 | 1.982 | 6.579 | 146.959 | 1.204 | 27.538 | 0.0 | 20.767 | 0.0 | 0.984 | 3.118 | 196.418 | 5.792 | 45.04 |
| 11 | 6.964 | 0.88 | 3.635 | 0.0 | 336.933 | 10.218 | 20.542 | 36.527 | 6.007 | 0.844 | 1.908 | 0.0 | 34.335 | 7.702 | 23.91 | 9.596 |
| 12 | 80.047 | 107.845 | 135.605 | 60.741 | 5.511 | 3.979 | 0.873 | 9.28 | 9.289 | 18.795 | 13.538 | 8.373 | 4.686 | 22.311 | 11.34 | 7.787 |
| 13 | 39.34 | 19.031 | 30.272 | 10.89 | 46.229 | 45.104 | 10.793 | 50.805 | 37.179 | 60.223 | 31.508 | 32.446 | 10.518 | 16.594 | 25.894 | 33.175 |
| 14 | 30.698 | 40.669 | 38.205 | 23.694 | 41.626 | 56.115 | 3.081 | 40.13 | 19.52 | 29.589 | 31.29 | 18.066 | 16.072 | 42.698 | 37.934 | 30.611 |
| 15 | 36.352 | 13.551 | 43.449 | 14.565 | 52.249 | 24.48 | 3.899 | 88.443 | 38.663 | 13.765 | 35.311 | 22.77 | 19.556 | 21.497 | 32.925 | 38.524 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.35 | -0.49 | 1.19 | -0.382 | -0.084 | -1.509 | -2.277 | 0.266 | -0.479 | -0.182 | 0.05 | -0.182 | -0.557 | -0.944 | 0.597 | 0.288 |
| 02 | -0.442 | -0.5 | 0.175 | 0.088 | -1.213 | -0.32 | -3.184 | -0.008 | -0.611 | 0.32 | 0.533 | 0.968 | -1.162 | 0.221 | -0.062 | 0.504 |
| 03 | -1.262 | -0.419 | -1.933 | -0.343 | -1.461 | 0.639 | -4.4 | 0.598 | -2.611 | 0.585 | -1.3 | 0.551 | -0.905 | 0.152 | -1.929 | 1.538 |
| 04 | -4.4 | -0.075 | -3.191 | -4.4 | -3.201 | 1.651 | -1.714 | -2.571 | -4.4 | -0.81 | -2.25 | -4.4 | -3.121 | 2.104 | -0.447 | -3.203 |
| 05 | -2.624 | -4.4 | -4.4 | -4.4 | 2.68 | -2.644 | -2.502 | -3.195 | -0.104 | -4.4 | -3.208 | -4.4 | -2.571 | -4.4 | -4.4 | -3.203 |
| 06 | 2.706 | -3.233 | -0.669 | -2.183 | -2.644 | -4.4 | -4.4 | -4.4 | -2.207 | -4.4 | -4.4 | -4.4 | -2.669 | -4.4 | -4.4 | -4.4 |
| 07 | -1.823 | -4.4 | 2.711 | -4.4 | -4.4 | -4.4 | -3.233 | -4.4 | -4.4 | -4.4 | -0.726 | -3.206 | -4.4 | -4.4 | -2.455 | -3.241 |
| 08 | -4.4 | -4.4 | -1.823 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.684 | -4.4 | 2.744 | -4.4 | -4.4 | -4.4 | -2.698 | -4.4 |
| 09 | -3.201 | -3.222 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 0.354 | 1.749 | -0.357 | 2.07 | -4.4 | -4.4 | -4.4 | -4.4 |
| 10 | -2.68 | 0.246 | -2.689 | -2.591 | -1.513 | 1.538 | -2.989 | -0.125 | -4.4 | -0.402 | -4.4 | -3.138 | -2.2 | 1.828 | -1.633 | 0.362 |
| 11 | -1.459 | -3.216 | -2.062 | -4.4 | 2.367 | -1.093 | -0.413 | 0.154 | -1.599 | -3.246 | -2.623 | -4.4 | 0.093 | -1.364 | -0.264 | -1.153 |
| 12 | 0.933 | 1.23 | 1.458 | 0.659 | -1.679 | -1.98 | -3.222 | -1.185 | -1.185 | -0.5 | -0.821 | -1.284 | -1.83 | -0.332 | -0.992 | -1.353 |
| 13 | 0.228 | -0.488 | -0.031 | -1.032 | 0.388 | 0.363 | -1.04 | 0.481 | 0.172 | 0.65 | 0.008 | 0.037 | -1.065 | -0.622 | -0.185 | 0.059 |
| 14 | -0.018 | 0.261 | 0.199 | -0.273 | 0.284 | 0.58 | -2.21 | 0.247 | -0.463 | -0.054 | 0.001 | -0.539 | -0.653 | 0.309 | 0.192 | -0.02 |
| 15 | 0.15 | -0.82 | 0.326 | -0.749 | 0.509 | -0.241 | -1.999 | 1.032 | 0.211 | -0.804 | 0.121 | -0.312 | -0.461 | -0.369 | 0.052 | 0.207 |