Model info
| Transcription factor | PRDM1 (GeneCards) | ||||||||
| Model | PRDM1_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 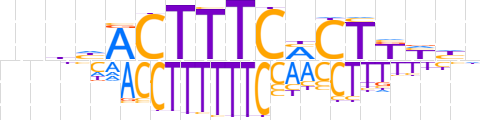 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 17 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ddRMAGKGAAAGTdhnn | ||||||||
| Best auROC (human) | 0.982 | ||||||||
| Best auROC (mouse) | 0.959 | ||||||||
| Peak sets in benchmark (human) | 12 | ||||||||
| Peak sets in benchmark (mouse) | 9 | ||||||||
| Aligned words | 501 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | PRDM1-like factors {2.3.3.12} | ||||||||
| HGNC | HGNC:9346 | ||||||||
| EntrezGene | GeneID:639 (SSTAR profile) | ||||||||
| UniProt ID | PRDM1_HUMAN | ||||||||
| UniProt AC | O75626 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | PRDM1 expression | ||||||||
| ReMap ChIP-seq dataset list | PRDM1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 57.76 | 11.61 | 90.91 | 17.213 | 39.708 | 9.532 | 7.955 | 12.338 | 44.911 | 7.216 | 32.45 | 8.599 | 40.591 | 9.295 | 93.494 | 16.419 |
| 02 | 117.166 | 12.231 | 28.624 | 24.949 | 10.721 | 14.137 | 2.067 | 10.728 | 175.855 | 12.508 | 24.334 | 12.111 | 11.956 | 10.118 | 17.125 | 15.371 |
| 03 | 265.567 | 10.966 | 28.036 | 11.13 | 24.727 | 10.181 | 1.935 | 12.15 | 45.305 | 15.365 | 6.83 | 4.65 | 23.104 | 21.786 | 12.473 | 5.795 |
| 04 | 319.883 | 0.0 | 32.507 | 6.313 | 57.513 | 0.0 | 0.784 | 0.0 | 38.472 | 0.788 | 5.505 | 4.511 | 26.434 | 0.0 | 4.587 | 2.704 |
| 05 | 26.865 | 4.194 | 400.309 | 10.932 | 0.0 | 0.0 | 0.788 | 0.0 | 0.94 | 0.0 | 42.443 | 0.0 | 1.378 | 0.0 | 7.79 | 4.359 |
| 06 | 8.732 | 0.0 | 6.931 | 13.521 | 0.839 | 1.053 | 0.0 | 2.301 | 67.189 | 9.317 | 74.59 | 300.235 | 6.669 | 0.771 | 3.073 | 4.778 |
| 07 | 0.902 | 1.174 | 81.353 | 0.0 | 0.902 | 0.0 | 10.24 | 0.0 | 2.504 | 0.0 | 82.09 | 0.0 | 13.141 | 0.895 | 306.8 | 0.0 |
| 08 | 16.73 | 0.0 | 0.0 | 0.718 | 2.069 | 0.0 | 0.0 | 0.0 | 478.941 | 0.0 | 0.0 | 1.542 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 476.62 | 0.0 | 18.265 | 2.855 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.26 | 0.0 | 0.0 | 0.0 |
| 10 | 469.162 | 0.0 | 4.34 | 5.378 | 0.0 | 0.0 | 0.0 | 0.0 | 18.265 | 0.0 | 0.0 | 0.0 | 1.955 | 0.0 | 0.899 | 0.0 |
| 11 | 13.785 | 7.175 | 461.666 | 6.756 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.406 | 0.833 | 0.0 | 0.0 | 4.129 | 1.249 |
| 12 | 1.784 | 2.53 | 3.53 | 5.941 | 0.0 | 3.332 | 1.068 | 2.776 | 5.744 | 31.35 | 18.989 | 414.117 | 0.0 | 0.739 | 2.52 | 5.579 |
| 13 | 0.701 | 0.0 | 3.099 | 3.728 | 18.959 | 3.014 | 5.762 | 10.216 | 11.231 | 1.753 | 9.123 | 4.0 | 85.968 | 30.776 | 222.527 | 89.141 |
| 14 | 44.818 | 25.519 | 26.12 | 20.402 | 20.15 | 9.817 | 0.0 | 5.577 | 132.069 | 46.126 | 38.501 | 23.817 | 24.712 | 15.069 | 23.636 | 43.669 |
| 15 | 87.523 | 23.761 | 53.504 | 56.96 | 30.651 | 14.727 | 6.564 | 44.589 | 20.515 | 19.193 | 22.54 | 26.008 | 21.152 | 15.481 | 24.633 | 32.198 |
| 16 | 55.1 | 15.705 | 51.455 | 37.581 | 21.915 | 18.359 | 3.449 | 29.44 | 40.649 | 18.109 | 24.118 | 24.365 | 13.354 | 33.222 | 44.837 | 68.342 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.609 | -0.97 | 1.06 | -0.586 | 0.237 | -1.16 | -1.333 | -0.911 | 0.359 | -1.426 | 0.037 | -1.259 | 0.259 | -1.184 | 1.088 | -0.633 |
| 02 | 1.313 | -0.919 | -0.087 | -0.222 | -1.047 | -0.778 | -2.556 | -1.046 | 1.717 | -0.897 | -0.247 | -0.929 | -0.941 | -1.102 | -0.591 | -0.697 |
| 03 | 2.129 | -1.025 | -0.107 | -1.01 | -0.231 | -1.096 | -2.611 | -0.926 | 0.368 | -0.697 | -1.478 | -1.837 | -0.298 | -0.355 | -0.9 | -1.632 |
| 04 | 2.315 | -4.4 | 0.039 | -1.552 | 0.604 | -4.4 | -3.295 | -4.4 | 0.206 | -3.292 | -1.681 | -1.865 | -0.165 | -4.4 | -1.85 | -2.325 |
| 05 | -0.149 | -1.932 | 2.539 | -1.028 | -4.4 | -4.4 | -3.292 | -4.4 | -3.17 | -4.4 | 0.303 | -4.4 | -2.885 | -4.4 | -1.353 | -1.897 |
| 06 | -1.244 | -4.4 | -1.464 | -0.822 | -3.249 | -3.088 | -4.4 | -2.465 | 0.759 | -1.182 | 0.863 | 2.251 | -1.5 | -3.306 | -2.213 | -1.812 |
| 07 | -3.2 | -3.008 | 0.949 | -4.4 | -3.2 | -4.4 | -1.091 | -4.4 | -2.392 | -4.4 | 0.958 | -4.4 | -0.849 | -3.205 | 2.273 | -4.4 |
| 08 | -0.614 | -4.4 | -4.4 | -3.353 | -2.555 | -4.4 | -4.4 | -4.4 | 2.718 | -4.4 | -4.4 | -2.797 | -4.4 | -4.4 | -4.4 | -4.4 |
| 09 | 2.713 | -4.4 | -0.528 | -2.278 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.48 | -4.4 | -4.4 | -4.4 |
| 10 | 2.697 | -4.4 | -1.901 | -1.702 | -4.4 | -4.4 | -4.4 | -4.4 | -0.528 | -4.4 | -4.4 | -4.4 | -2.603 | -4.4 | -3.201 | -4.4 |
| 11 | -0.803 | -1.431 | 2.681 | -1.488 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -1.887 | -3.254 | -4.4 | -4.4 | -1.946 | -2.961 |
| 12 | -2.679 | -2.383 | -2.089 | -1.609 | -4.4 | -2.14 | -3.079 | -2.302 | -1.641 | 0.003 | -0.49 | 2.573 | -4.4 | -3.334 | -2.387 | -1.668 |
| 13 | -3.369 | -4.4 | -2.205 | -2.039 | -0.492 | -2.23 | -1.638 | -1.093 | -1.002 | -2.693 | -1.202 | -1.975 | 1.004 | -0.015 | 1.952 | 1.04 |
| 14 | 0.357 | -0.2 | -0.177 | -0.42 | -0.432 | -1.131 | -4.4 | -1.668 | 1.432 | 0.385 | 0.206 | -0.268 | -0.232 | -0.716 | -0.275 | 0.331 |
| 15 | 1.022 | -0.27 | 0.533 | 0.595 | -0.019 | -0.739 | -1.515 | 0.352 | -0.414 | -0.48 | -0.322 | -0.181 | -0.384 | -0.69 | -0.235 | 0.03 |
| 16 | 0.562 | -0.676 | 0.494 | 0.182 | -0.35 | -0.523 | -2.11 | -0.059 | 0.26 | -0.537 | -0.255 | -0.245 | -0.834 | 0.06 | 0.357 | 0.776 |