Model info
| Transcription factor | SMAD2 (GeneCards) | ||||||||
| Model | SMAD2_HUMAN.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 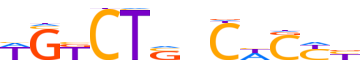 | ||||||||
| LOGO (reverse complement) | 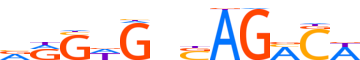 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | WGKCTSnChCYb | ||||||||
| Best auROC (human) | 0.848 | ||||||||
| Best auROC (mouse) | 0.808 | ||||||||
| Peak sets in benchmark (human) | 17 | ||||||||
| Peak sets in benchmark (mouse) | 26 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | SMAD factors {7.1.1} | ||||||||
| TF subfamily | Regulatory Smads (R-Smad) {7.1.1.1} | ||||||||
| HGNC | HGNC:6768 | ||||||||
| EntrezGene | GeneID:4087 (SSTAR profile) | ||||||||
| UniProt ID | SMAD2_HUMAN | ||||||||
| UniProt AC | Q15796 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | SMAD2 expression | ||||||||
| ReMap ChIP-seq dataset list | SMAD2 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 107.0 | 37.0 | 24.0 | 332.0 |
| 02 | 22.0 | 37.0 | 427.0 | 14.0 |
| 03 | 26.0 | 60.0 | 73.0 | 341.0 |
| 04 | 2.0 | 483.0 | 4.0 | 11.0 |
| 05 | 10.0 | 10.0 | 0.0 | 480.0 |
| 06 | 61.0 | 108.0 | 302.0 | 29.0 |
| 07 | 81.0 | 134.0 | 125.0 | 160.0 |
| 08 | 16.0 | 450.0 | 14.0 | 20.0 |
| 09 | 268.0 | 59.0 | 27.0 | 146.0 |
| 10 | 24.0 | 402.0 | 55.0 | 19.0 |
| 11 | 80.0 | 302.0 | 4.0 | 114.0 |
| 12 | 46.0 | 131.0 | 54.0 | 269.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.153 | -1.189 | -1.6 | 0.969 |
| 02 | -1.681 | -1.189 | 1.22 | -2.096 |
| 03 | -1.525 | -0.721 | -0.529 | 0.996 |
| 04 | -3.573 | 1.343 | -3.126 | -2.311 |
| 05 | -2.394 | -2.394 | -4.4 | 1.336 |
| 06 | -0.705 | -0.144 | 0.875 | -1.421 |
| 07 | -0.427 | 0.069 | 0.0 | 0.244 |
| 08 | -1.975 | 1.272 | -2.096 | -1.77 |
| 09 | 0.756 | -0.737 | -1.489 | 0.154 |
| 10 | -1.6 | 1.16 | -0.805 | -1.818 |
| 11 | -0.439 | 0.875 | -3.126 | -0.091 |
| 12 | -0.979 | 0.046 | -0.823 | 0.76 |