Model info
| Transcription factor | Sox2 | ||||||||
| Model | SOX2_MOUSE.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 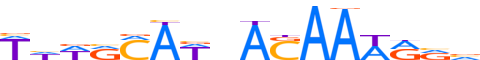 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bbYWTTSTnWTKYWdW | ||||||||
| Best auROC (human) | 0.955 | ||||||||
| Best auROC (mouse) | 0.958 | ||||||||
| Peak sets in benchmark (human) | 67 | ||||||||
| Peak sets in benchmark (mouse) | 109 | ||||||||
| Aligned words | 499 | ||||||||
| TF family | SOX-related factors {4.1.1} | ||||||||
| TF subfamily | Group B {4.1.1.2} | ||||||||
| MGI | MGI:98364 | ||||||||
| EntrezGene | GeneID:20674 (SSTAR profile) | ||||||||
| UniProt ID | SOX2_MOUSE | ||||||||
| UniProt AC | P48432 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Sox2 expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 35.0 | 92.0 | 146.0 | 226.0 |
| 02 | 28.0 | 281.0 | 71.0 | 119.0 |
| 03 | 25.0 | 272.0 | 8.0 | 194.0 |
| 04 | 219.0 | 6.0 | 4.0 | 270.0 |
| 05 | 1.0 | 11.0 | 5.0 | 482.0 |
| 06 | 7.0 | 1.0 | 2.0 | 489.0 |
| 07 | 39.0 | 76.0 | 372.0 | 12.0 |
| 08 | 52.0 | 1.0 | 12.0 | 434.0 |
| 09 | 113.0 | 124.0 | 78.0 | 184.0 |
| 10 | 340.0 | 39.0 | 33.0 | 87.0 |
| 11 | 36.0 | 5.0 | 19.0 | 439.0 |
| 12 | 19.0 | 29.0 | 375.0 | 76.0 |
| 13 | 24.0 | 317.0 | 59.0 | 99.0 |
| 14 | 279.0 | 51.0 | 17.0 | 152.0 |
| 15 | 277.0 | 47.0 | 72.0 | 103.0 |
| 16 | 386.0 | 19.0 | 38.0 | 56.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -1.24 | -0.3 | 0.156 | 0.589 |
| 02 | -1.452 | 0.805 | -0.554 | -0.047 |
| 03 | -1.56 | 0.773 | -2.582 | 0.437 |
| 04 | 0.557 | -2.817 | -3.124 | 0.765 |
| 05 | -3.901 | -2.309 | -2.959 | 1.342 |
| 06 | -2.692 | -3.901 | -3.571 | 1.357 |
| 07 | -1.136 | -0.488 | 1.084 | -2.232 |
| 08 | -0.858 | -3.901 | -2.232 | 1.238 |
| 09 | -0.098 | -0.006 | -0.462 | 0.385 |
| 10 | 0.995 | -1.136 | -1.296 | -0.355 |
| 11 | -1.213 | -2.959 | -1.816 | 1.249 |
| 12 | -1.816 | -1.419 | 1.092 | -0.488 |
| 13 | -1.598 | 0.925 | -0.735 | -0.228 |
| 14 | 0.798 | -0.877 | -1.918 | 0.195 |
| 15 | 0.791 | -0.956 | -0.541 | -0.189 |
| 16 | 1.121 | -1.816 | -1.161 | -0.786 |