Model info
| Transcription factor | Stat6 | ||||||||
| Model | STAT6_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 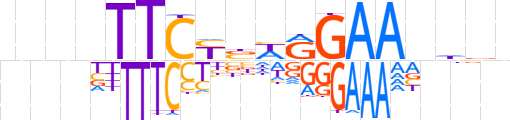 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbdnTTCYWvRGAAnnnn | ||||||||
| Best auROC (human) | 0.771 | ||||||||
| Best auROC (mouse) | 0.951 | ||||||||
| Peak sets in benchmark (human) | 19 | ||||||||
| Peak sets in benchmark (mouse) | 24 | ||||||||
| Aligned words | 492 | ||||||||
| TF family | STAT factors {6.2.1} | ||||||||
| TF subfamily | STAT6 {6.2.1.0.7} | ||||||||
| MGI | MGI:103034 | ||||||||
| EntrezGene | GeneID:20852 (SSTAR profile) | ||||||||
| UniProt ID | STAT6_MOUSE | ||||||||
| UniProt AC | P52633 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Stat6 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 12.0 | 31.0 | 47.0 | 7.0 | 28.0 | 45.0 | 14.0 | 37.0 | 16.0 | 49.0 | 38.0 | 24.0 | 8.0 | 52.0 | 47.0 | 29.0 |
| 02 | 34.0 | 5.0 | 17.0 | 8.0 | 97.0 | 25.0 | 7.0 | 48.0 | 72.0 | 22.0 | 29.0 | 23.0 | 29.0 | 19.0 | 24.0 | 25.0 |
| 03 | 48.0 | 33.0 | 75.0 | 76.0 | 13.0 | 20.0 | 2.0 | 36.0 | 21.0 | 20.0 | 11.0 | 25.0 | 12.0 | 33.0 | 16.0 | 43.0 |
| 04 | 1.0 | 0.0 | 1.0 | 92.0 | 0.0 | 0.0 | 0.0 | 106.0 | 0.0 | 3.0 | 0.0 | 101.0 | 0.0 | 0.0 | 0.0 | 180.0 |
| 05 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 4.0 | 474.0 |
| 06 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 24.0 | 445.0 | 2.0 | 8.0 |
| 07 | 8.0 | 15.0 | 0.0 | 1.0 | 18.0 | 300.0 | 0.0 | 132.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 7.0 | 0.0 | 1.0 |
| 08 | 16.0 | 5.0 | 1.0 | 4.0 | 150.0 | 4.0 | 8.0 | 161.0 | 0.0 | 0.0 | 0.0 | 0.0 | 36.0 | 0.0 | 37.0 | 62.0 |
| 09 | 58.0 | 54.0 | 78.0 | 12.0 | 0.0 | 2.0 | 7.0 | 0.0 | 0.0 | 15.0 | 31.0 | 0.0 | 56.0 | 64.0 | 107.0 | 0.0 |
| 10 | 60.0 | 1.0 | 48.0 | 5.0 | 105.0 | 0.0 | 14.0 | 16.0 | 114.0 | 5.0 | 93.0 | 11.0 | 4.0 | 0.0 | 6.0 | 2.0 |
| 11 | 2.0 | 0.0 | 272.0 | 9.0 | 1.0 | 0.0 | 3.0 | 2.0 | 24.0 | 0.0 | 135.0 | 2.0 | 0.0 | 0.0 | 33.0 | 1.0 |
| 12 | 27.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 443.0 | 0.0 | 0.0 | 0.0 | 14.0 | 0.0 | 0.0 | 0.0 |
| 13 | 478.0 | 2.0 | 4.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 176.0 | 105.0 | 110.0 | 87.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 15 | 53.0 | 43.0 | 44.0 | 37.0 | 25.0 | 22.0 | 6.0 | 56.0 | 20.0 | 29.0 | 14.0 | 47.0 | 6.0 | 22.0 | 17.0 | 43.0 |
| 16 | 17.0 | 32.0 | 44.0 | 11.0 | 28.0 | 47.0 | 5.0 | 36.0 | 13.0 | 33.0 | 25.0 | 10.0 | 14.0 | 53.0 | 68.0 | 48.0 |
| 17 | 9.0 | 17.0 | 38.0 | 8.0 | 62.0 | 49.0 | 8.0 | 46.0 | 36.0 | 32.0 | 57.0 | 17.0 | 13.0 | 26.0 | 35.0 | 31.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.906 | 0.024 | 0.436 | -1.423 | -0.076 | 0.393 | -0.756 | 0.199 | -0.626 | 0.477 | 0.226 | -0.228 | -1.296 | 0.536 | 0.436 | -0.042 |
| 02 | 0.115 | -1.738 | -0.567 | -1.296 | 1.156 | -0.188 | -1.423 | 0.457 | 0.86 | -0.314 | -0.042 | -0.27 | -0.042 | -0.458 | -0.228 | -0.188 |
| 03 | 0.457 | 0.086 | 0.9 | 0.914 | -0.828 | -0.407 | -2.552 | 0.172 | -0.359 | -0.407 | -0.99 | -0.188 | -0.906 | 0.086 | -0.626 | 0.348 |
| 04 | -3.095 | -4.373 | -3.095 | 1.104 | -4.373 | -4.373 | -4.373 | 1.245 | -4.373 | -2.202 | -4.373 | 1.197 | -4.373 | -4.373 | -4.373 | 1.773 |
| 05 | -4.373 | -4.373 | -4.373 | -3.095 | -4.373 | -4.373 | -4.373 | -2.202 | -4.373 | -4.373 | -4.373 | -3.095 | -3.095 | -4.373 | -1.944 | 2.74 |
| 06 | -4.373 | -3.095 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -1.944 | -4.373 | -4.373 | -0.228 | 2.677 | -2.552 | -1.296 |
| 07 | -1.296 | -0.689 | -4.373 | -3.095 | -0.511 | 2.283 | -4.373 | 1.464 | -4.373 | -3.095 | -4.373 | -3.095 | -4.373 | -1.423 | -4.373 | -3.095 |
| 08 | -0.626 | -1.738 | -3.095 | -1.944 | 1.591 | -1.944 | -1.296 | 1.662 | -4.373 | -4.373 | -4.373 | -4.373 | 0.172 | -4.373 | 0.199 | 0.711 |
| 09 | 0.645 | 0.574 | 0.939 | -0.906 | -4.373 | -2.552 | -1.423 | -4.373 | -4.373 | -0.689 | 0.024 | -4.373 | 0.61 | 0.743 | 1.254 | -4.373 |
| 10 | 0.679 | -3.095 | 0.457 | -1.738 | 1.235 | -4.373 | -0.756 | -0.626 | 1.317 | -1.738 | 1.115 | -0.99 | -1.944 | -4.373 | -1.568 | -2.552 |
| 11 | -2.552 | -4.373 | 2.185 | -1.183 | -3.095 | -4.373 | -2.202 | -2.552 | -0.228 | -4.373 | 1.486 | -2.552 | -4.373 | -4.373 | 0.086 | -3.095 |
| 12 | -0.112 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | 2.672 | -4.373 | -4.373 | -4.373 | -0.756 | -4.373 | -4.373 | -4.373 |
| 13 | 2.748 | -2.552 | -1.944 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 |
| 14 | 1.75 | 1.235 | 1.282 | 1.048 | -4.373 | -3.095 | -4.373 | -3.095 | -3.095 | -2.202 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 |
| 15 | 0.555 | 0.348 | 0.371 | 0.199 | -0.188 | -0.314 | -1.568 | 0.61 | -0.407 | -0.042 | -0.756 | 0.436 | -1.568 | -0.314 | -0.567 | 0.348 |
| 16 | -0.567 | 0.056 | 0.371 | -0.99 | -0.076 | 0.436 | -1.738 | 0.172 | -0.828 | 0.086 | -0.188 | -1.082 | -0.756 | 0.555 | 0.803 | 0.457 |
| 17 | -1.183 | -0.567 | 0.226 | -1.296 | 0.711 | 0.477 | -1.296 | 0.415 | 0.172 | 0.056 | 0.628 | -0.567 | -0.828 | -0.149 | 0.144 | 0.024 |