Model info
| Transcription factor | TLX1 (GeneCards) | ||||||||
| Model | TLX1_HUMAN.H11MO.0.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 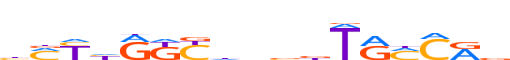 | ||||||||
| Data source | Integrative | ||||||||
| Model release | HOCOMOCOv9 | ||||||||
| Model length | 17 | ||||||||
| Quality | D | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | hYKKYAvbndvMYnRKd | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 50 | ||||||||
| TF family | NK-related factors {3.1.2} | ||||||||
| TF subfamily | TLX {3.1.2.21} | ||||||||
| HGNC | HGNC:5056 | ||||||||
| EntrezGene | GeneID:3195 (SSTAR profile) | ||||||||
| UniProt ID | TLX1_HUMAN | ||||||||
| UniProt AC | P31314 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | TLX1 expression | ||||||||
| ReMap ChIP-seq dataset list | TLX1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 9.236 | 21.151 | 7.605 | 12.008 |
| 02 | 1.631 | 17.946 | 2.538 | 27.884 |
| 03 | 0.816 | 1.631 | 36.132 | 11.42 |
| 04 | 7.342 | 3.263 | 30.332 | 9.064 |
| 05 | 0.816 | 27.884 | 0.816 | 20.484 |
| 06 | 42.334 | 1.386 | 0.816 | 5.465 |
| 07 | 19.177 | 17.613 | 7.745 | 5.465 |
| 08 | 5.219 | 15.982 | 16.148 | 12.651 |
| 09 | 13.78 | 7.096 | 12.64 | 16.484 |
| 10 | 14.517 | 4.973 | 13.467 | 17.043 |
| 11 | 8.157 | 8.157 | 33.685 | 0.0 |
| 12 | 11.42 | 31.238 | 0.0 | 7.342 |
| 13 | 2.447 | 33.595 | 0.0 | 13.958 |
| 14 | 25.437 | 6.617 | 8.973 | 8.973 |
| 15 | 33.27 | 0.816 | 8.728 | 7.187 |
| 16 | 3.588 | 5.219 | 29.94 | 11.254 |
| 17 | 14.126 | 6.627 | 20.418 | 8.828 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.277 | 0.496 | -0.451 | -0.037 |
| 02 | -1.642 | 0.339 | -1.344 | 0.761 |
| 03 | -2.017 | -1.642 | 1.013 | -0.083 |
| 04 | -0.482 | -1.156 | 0.843 | -0.294 |
| 05 | -2.017 | 0.761 | -2.017 | 0.465 |
| 06 | 1.167 | -1.741 | -2.017 | -0.738 |
| 07 | 0.402 | 0.322 | -0.435 | -0.738 |
| 08 | -0.777 | 0.23 | 0.24 | 0.011 |
| 09 | 0.091 | -0.512 | 0.01 | 0.259 |
| 10 | 0.139 | -0.817 | 0.069 | 0.29 |
| 11 | -0.389 | -0.389 | 0.945 | -2.623 |
| 12 | -0.083 | 0.871 | -2.623 | -0.482 |
| 13 | -1.37 | 0.942 | -2.623 | 0.103 |
| 14 | 0.673 | -0.574 | -0.303 | -0.303 |
| 15 | 0.933 | -2.017 | -0.328 | -0.501 |
| 16 | -1.083 | -0.777 | 0.83 | -0.097 |
| 17 | 0.114 | -0.572 | 0.462 | -0.318 |