Model info
| Transcription factor | ZNF708 (GeneCards) | ||||||||
| Model | ZN708_HUMAN.H11MO.0.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 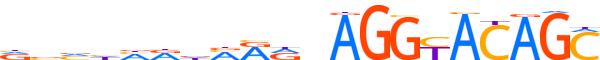 | ||||||||
| LOGO (reverse complement) | 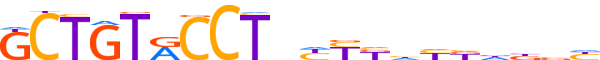 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 20 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vvvhhddMRRnAGGYACAGC | ||||||||
| Best auROC (human) | 0.838 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 359 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:12945 | ||||||||
| EntrezGene | GeneID:7562 (SSTAR profile) | ||||||||
| UniProt ID | ZN708_HUMAN | ||||||||
| UniProt AC | P17019 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF708 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF708 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 61.0 | 48.0 | 208.0 | 42.0 |
| 02 | 74.0 | 161.0 | 79.0 | 45.0 |
| 03 | 85.0 | 179.0 | 54.0 | 41.0 |
| 04 | 76.0 | 68.0 | 39.0 | 176.0 |
| 05 | 209.0 | 60.0 | 43.0 | 47.0 |
| 06 | 201.0 | 27.0 | 88.0 | 43.0 |
| 07 | 73.0 | 47.0 | 72.0 | 167.0 |
| 08 | 215.0 | 61.0 | 54.0 | 29.0 |
| 09 | 233.0 | 30.0 | 85.0 | 11.0 |
| 10 | 99.0 | 13.0 | 196.0 | 51.0 |
| 11 | 55.0 | 112.0 | 93.0 | 99.0 |
| 12 | 336.0 | 0.0 | 15.0 | 8.0 |
| 13 | 3.0 | 1.0 | 353.0 | 2.0 |
| 14 | 6.0 | 15.0 | 334.0 | 4.0 |
| 15 | 13.0 | 151.0 | 2.0 | 193.0 |
| 16 | 339.0 | 8.0 | 9.0 | 3.0 |
| 17 | 2.0 | 313.0 | 7.0 | 37.0 |
| 18 | 331.0 | 2.0 | 22.0 | 4.0 |
| 19 | 14.0 | 2.0 | 340.0 | 3.0 |
| 20 | 30.0 | 300.0 | 9.0 | 20.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.379 | -0.612 | 0.831 | -0.741 |
| 02 | -0.19 | 0.577 | -0.125 | -0.674 |
| 03 | -0.053 | 0.682 | -0.497 | -0.764 |
| 04 | -0.163 | -0.272 | -0.813 | 0.666 |
| 05 | 0.836 | -0.395 | -0.718 | -0.632 |
| 06 | 0.797 | -1.164 | -0.019 | -0.718 |
| 07 | -0.203 | -0.632 | -0.216 | 0.613 |
| 08 | 0.864 | -0.379 | -0.497 | -1.097 |
| 09 | 0.944 | -1.064 | -0.053 | -1.99 |
| 10 | 0.097 | -1.841 | 0.772 | -0.553 |
| 11 | -0.48 | 0.218 | 0.035 | 0.097 |
| 12 | 1.308 | -4.127 | -1.712 | -2.265 |
| 13 | -3.016 | -3.609 | 1.357 | -3.269 |
| 14 | -2.502 | -1.712 | 1.302 | -2.814 |
| 15 | -1.841 | 0.514 | -3.269 | 0.757 |
| 16 | 1.317 | -2.265 | -2.165 | -3.016 |
| 17 | -3.269 | 1.238 | -2.377 | -0.863 |
| 18 | 1.293 | -3.269 | -1.358 | -2.814 |
| 19 | -1.774 | -3.269 | 1.32 | -3.016 |
| 20 | -1.064 | 1.195 | -2.165 | -1.447 |