Model info
| Transcription factor | MYNN (GeneCards) | ||||||||
| Model | MYNN_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 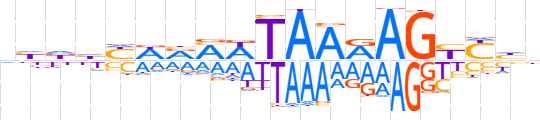 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndRRCTYTTAWKYWRdhdn | ||||||||
| Best auROC (human) | 0.82 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 4 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 389 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:14955 | ||||||||
| EntrezGene | GeneID:55892 (SSTAR profile) | ||||||||
| UniProt ID | MYNN_HUMAN | ||||||||
| UniProt AC | Q9NPC7 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | MYNN expression | ||||||||
| ReMap ChIP-seq dataset list | MYNN datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 26.0 | 3.0 | 64.0 | 4.0 | 36.0 | 10.0 | 12.0 | 19.0 | 39.0 | 9.0 | 66.0 | 23.0 | 15.0 | 8.0 | 39.0 | 11.0 |
| 02 | 14.0 | 5.0 | 87.0 | 10.0 | 8.0 | 3.0 | 11.0 | 8.0 | 29.0 | 8.0 | 131.0 | 13.0 | 3.0 | 3.0 | 46.0 | 5.0 |
| 03 | 16.0 | 0.0 | 37.0 | 1.0 | 10.0 | 2.0 | 6.0 | 1.0 | 169.0 | 12.0 | 78.0 | 16.0 | 7.0 | 2.0 | 24.0 | 3.0 |
| 04 | 3.0 | 196.0 | 2.0 | 1.0 | 2.0 | 13.0 | 1.0 | 0.0 | 2.0 | 139.0 | 3.0 | 1.0 | 1.0 | 17.0 | 3.0 | 0.0 |
| 05 | 0.0 | 1.0 | 1.0 | 6.0 | 1.0 | 4.0 | 1.0 | 359.0 | 0.0 | 4.0 | 2.0 | 3.0 | 0.0 | 0.0 | 1.0 | 1.0 |
| 06 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 8.0 | 1.0 | 0.0 | 0.0 | 4.0 | 13.0 | 124.0 | 11.0 | 221.0 |
| 07 | 3.0 | 3.0 | 2.0 | 6.0 | 1.0 | 5.0 | 9.0 | 110.0 | 0.0 | 4.0 | 2.0 | 5.0 | 4.0 | 9.0 | 10.0 | 211.0 |
| 08 | 0.0 | 0.0 | 0.0 | 8.0 | 2.0 | 0.0 | 0.0 | 19.0 | 10.0 | 0.0 | 0.0 | 13.0 | 2.0 | 3.0 | 1.0 | 326.0 |
| 09 | 12.0 | 0.0 | 2.0 | 0.0 | 2.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 334.0 | 9.0 | 20.0 | 3.0 |
| 10 | 90.0 | 33.0 | 6.0 | 220.0 | 0.0 | 2.0 | 0.0 | 8.0 | 1.0 | 5.0 | 1.0 | 15.0 | 0.0 | 0.0 | 1.0 | 2.0 |
| 11 | 6.0 | 22.0 | 21.0 | 42.0 | 4.0 | 5.0 | 7.0 | 24.0 | 1.0 | 1.0 | 0.0 | 6.0 | 3.0 | 21.0 | 28.0 | 193.0 |
| 12 | 3.0 | 4.0 | 4.0 | 3.0 | 8.0 | 10.0 | 3.0 | 28.0 | 7.0 | 10.0 | 16.0 | 23.0 | 24.0 | 39.0 | 21.0 | 181.0 |
| 13 | 2.0 | 5.0 | 5.0 | 30.0 | 16.0 | 10.0 | 1.0 | 36.0 | 19.0 | 5.0 | 3.0 | 17.0 | 21.0 | 36.0 | 14.0 | 164.0 |
| 14 | 19.0 | 8.0 | 25.0 | 6.0 | 22.0 | 6.0 | 13.0 | 15.0 | 3.0 | 5.0 | 10.0 | 5.0 | 11.0 | 11.0 | 202.0 | 23.0 |
| 15 | 15.0 | 1.0 | 17.0 | 22.0 | 12.0 | 8.0 | 0.0 | 10.0 | 153.0 | 20.0 | 49.0 | 28.0 | 7.0 | 11.0 | 16.0 | 15.0 |
| 16 | 130.0 | 31.0 | 4.0 | 22.0 | 16.0 | 11.0 | 3.0 | 10.0 | 34.0 | 18.0 | 11.0 | 19.0 | 14.0 | 27.0 | 14.0 | 20.0 |
| 17 | 107.0 | 12.0 | 61.0 | 14.0 | 55.0 | 8.0 | 7.0 | 17.0 | 13.0 | 7.0 | 8.0 | 4.0 | 28.0 | 10.0 | 12.0 | 21.0 |
| 18 | 52.0 | 30.0 | 50.0 | 71.0 | 9.0 | 13.0 | 3.0 | 12.0 | 15.0 | 26.0 | 26.0 | 21.0 | 12.0 | 15.0 | 9.0 | 20.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.079 | -1.978 | 0.971 | -1.718 | 0.4 | -0.854 | -0.678 | -0.23 | 0.48 | -0.956 | 1.002 | -0.042 | -0.461 | -1.069 | 0.48 | -0.762 |
| 02 | -0.528 | -1.512 | 1.277 | -0.854 | -1.069 | -1.978 | -0.762 | -1.069 | 0.187 | -1.069 | 1.685 | -0.6 | -1.978 | -1.978 | 0.643 | -1.512 |
| 03 | -0.398 | -4.183 | 0.427 | -2.877 | -0.854 | -2.33 | -1.342 | -2.877 | 1.939 | -0.678 | 1.168 | -0.398 | -1.196 | -2.33 | 0.0 | -1.978 |
| 04 | -1.978 | 2.087 | -2.33 | -2.877 | -2.33 | -0.6 | -2.877 | -4.183 | -2.33 | 1.744 | -1.978 | -2.877 | -2.877 | -0.339 | -1.978 | -4.183 |
| 05 | -4.183 | -2.877 | -2.877 | -1.342 | -2.877 | -1.718 | -2.877 | 2.691 | -4.183 | -1.718 | -2.33 | -1.978 | -4.183 | -4.183 | -2.877 | -2.877 |
| 06 | -4.183 | -4.183 | -4.183 | -2.877 | -4.183 | -2.877 | -4.183 | -1.069 | -2.877 | -4.183 | -4.183 | -1.718 | -0.6 | 1.63 | -0.762 | 2.206 |
| 07 | -1.978 | -1.978 | -2.33 | -1.342 | -2.877 | -1.512 | -0.956 | 1.51 | -4.183 | -1.718 | -2.33 | -1.512 | -1.718 | -0.956 | -0.854 | 2.16 |
| 08 | -4.183 | -4.183 | -4.183 | -1.069 | -2.33 | -4.183 | -4.183 | -0.23 | -0.854 | -4.183 | -4.183 | -0.6 | -2.33 | -1.978 | -2.877 | 2.595 |
| 09 | -0.678 | -4.183 | -2.33 | -4.183 | -2.33 | -2.877 | -4.183 | -4.183 | -2.877 | -4.183 | -4.183 | -4.183 | 2.619 | -0.956 | -0.179 | -1.978 |
| 10 | 1.311 | 0.314 | -1.342 | 2.202 | -4.183 | -2.33 | -4.183 | -1.069 | -2.877 | -1.512 | -2.877 | -0.461 | -4.183 | -4.183 | -2.877 | -2.33 |
| 11 | -1.342 | -0.086 | -0.131 | 0.553 | -1.718 | -1.512 | -1.196 | 0.0 | -2.877 | -2.877 | -4.183 | -1.342 | -1.978 | -0.131 | 0.152 | 2.071 |
| 12 | -1.978 | -1.718 | -1.718 | -1.978 | -1.069 | -0.854 | -1.978 | 0.152 | -1.196 | -0.854 | -0.398 | -0.042 | 0.0 | 0.48 | -0.131 | 2.007 |
| 13 | -2.33 | -1.512 | -1.512 | 0.22 | -0.398 | -0.854 | -2.877 | 0.4 | -0.23 | -1.512 | -1.978 | -0.339 | -0.131 | 0.4 | -0.528 | 1.909 |
| 14 | -0.23 | -1.069 | 0.04 | -1.342 | -0.086 | -1.342 | -0.6 | -0.461 | -1.978 | -1.512 | -0.854 | -1.512 | -0.762 | -0.762 | 2.117 | -0.042 |
| 15 | -0.461 | -2.877 | -0.339 | -0.086 | -0.678 | -1.069 | -4.183 | -0.854 | 1.839 | -0.179 | 0.706 | 0.152 | -1.196 | -0.762 | -0.398 | -0.461 |
| 16 | 1.677 | 0.252 | -1.718 | -0.086 | -0.398 | -0.762 | -1.978 | -0.854 | 0.344 | -0.283 | -0.762 | -0.23 | -0.528 | 0.116 | -0.528 | -0.179 |
| 17 | 1.483 | -0.678 | 0.924 | -0.528 | 0.821 | -1.069 | -1.196 | -0.339 | -0.6 | -1.196 | -1.069 | -1.718 | 0.152 | -0.854 | -0.678 | -0.131 |
| 18 | 0.765 | 0.22 | 0.726 | 1.074 | -0.956 | -0.6 | -1.978 | -0.678 | -0.461 | 0.079 | 0.079 | -0.131 | -0.678 | -0.461 | -0.956 | -0.179 |