Model info
| Transcription factor | VDR (GeneCards) | ||||||||
| Model | VDR_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 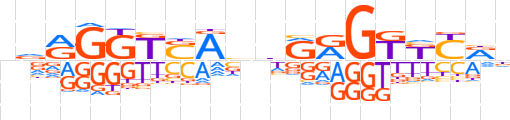 | ||||||||
| LOGO (reverse complement) | 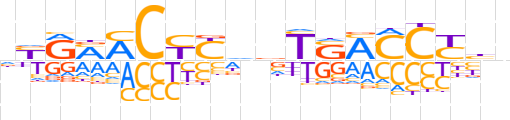 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvRGGKSAbhRRGKKYRn | ||||||||
| Best auROC (human) | 0.95 | ||||||||
| Best auROC (mouse) | 0.939 | ||||||||
| Peak sets in benchmark (human) | 48 | ||||||||
| Peak sets in benchmark (mouse) | 30 | ||||||||
| Aligned words | 419 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1) {2.1.2} | ||||||||
| TF subfamily | Vitamin D receptor (NR1I) {2.1.2.4} | ||||||||
| HGNC | HGNC:12679 | ||||||||
| EntrezGene | GeneID:7421 (SSTAR profile) | ||||||||
| UniProt ID | VDR_HUMAN | ||||||||
| UniProt AC | P11473 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | VDR expression | ||||||||
| ReMap ChIP-seq dataset list | VDR datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 40.0 | 11.0 | 69.0 | 11.0 | 50.0 | 21.0 | 26.0 | 18.0 | 25.0 | 16.0 | 33.0 | 6.0 | 14.0 | 10.0 | 61.0 | 8.0 |
| 02 | 51.0 | 3.0 | 75.0 | 0.0 | 42.0 | 4.0 | 10.0 | 2.0 | 91.0 | 3.0 | 94.0 | 1.0 | 15.0 | 0.0 | 28.0 | 0.0 |
| 03 | 16.0 | 1.0 | 179.0 | 3.0 | 7.0 | 0.0 | 1.0 | 2.0 | 26.0 | 3.0 | 172.0 | 6.0 | 0.0 | 0.0 | 3.0 | 0.0 |
| 04 | 4.0 | 0.0 | 43.0 | 2.0 | 3.0 | 0.0 | 0.0 | 1.0 | 10.0 | 0.0 | 268.0 | 77.0 | 1.0 | 0.0 | 9.0 | 1.0 |
| 05 | 0.0 | 2.0 | 2.0 | 14.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.0 | 23.0 | 49.0 | 242.0 | 1.0 | 3.0 | 16.0 | 61.0 |
| 06 | 3.0 | 1.0 | 2.0 | 1.0 | 5.0 | 21.0 | 0.0 | 2.0 | 3.0 | 55.0 | 6.0 | 3.0 | 10.0 | 209.0 | 69.0 | 29.0 |
| 07 | 11.0 | 0.0 | 7.0 | 3.0 | 248.0 | 5.0 | 19.0 | 14.0 | 52.0 | 6.0 | 16.0 | 3.0 | 27.0 | 2.0 | 5.0 | 1.0 |
| 08 | 53.0 | 120.0 | 102.0 | 63.0 | 2.0 | 7.0 | 1.0 | 3.0 | 6.0 | 22.0 | 12.0 | 7.0 | 0.0 | 10.0 | 2.0 | 9.0 |
| 09 | 20.0 | 13.0 | 5.0 | 23.0 | 41.0 | 28.0 | 9.0 | 81.0 | 17.0 | 32.0 | 28.0 | 40.0 | 13.0 | 15.0 | 22.0 | 32.0 |
| 10 | 3.0 | 7.0 | 80.0 | 1.0 | 31.0 | 10.0 | 39.0 | 8.0 | 8.0 | 10.0 | 45.0 | 1.0 | 10.0 | 19.0 | 146.0 | 1.0 |
| 11 | 23.0 | 1.0 | 28.0 | 0.0 | 34.0 | 0.0 | 10.0 | 2.0 | 148.0 | 3.0 | 153.0 | 6.0 | 2.0 | 0.0 | 9.0 | 0.0 |
| 12 | 0.0 | 0.0 | 207.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 5.0 | 0.0 | 191.0 | 4.0 | 0.0 | 0.0 | 8.0 | 0.0 |
| 13 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 7.0 | 136.0 | 265.0 | 0.0 | 0.0 | 3.0 | 1.0 |
| 14 | 1.0 | 0.0 | 1.0 | 0.0 | 2.0 | 2.0 | 1.0 | 2.0 | 20.0 | 19.0 | 29.0 | 76.0 | 13.0 | 19.0 | 31.0 | 203.0 |
| 15 | 0.0 | 32.0 | 2.0 | 2.0 | 1.0 | 26.0 | 1.0 | 12.0 | 1.0 | 47.0 | 6.0 | 8.0 | 0.0 | 193.0 | 16.0 | 72.0 |
| 16 | 0.0 | 0.0 | 2.0 | 0.0 | 223.0 | 16.0 | 30.0 | 29.0 | 15.0 | 5.0 | 2.0 | 3.0 | 26.0 | 20.0 | 39.0 | 9.0 |
| 17 | 54.0 | 60.0 | 68.0 | 82.0 | 11.0 | 12.0 | 3.0 | 15.0 | 13.0 | 11.0 | 12.0 | 37.0 | 8.0 | 12.0 | 6.0 | 15.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.419 | -0.848 | 0.96 | -0.848 | 0.64 | -0.217 | -0.007 | -0.368 | -0.046 | -0.484 | 0.228 | -1.427 | -0.614 | -0.94 | 0.837 | -1.154 |
| 02 | 0.66 | -2.062 | 1.043 | -4.254 | 0.467 | -1.803 | -0.94 | -2.414 | 1.235 | -2.062 | 1.268 | -2.959 | -0.547 | -4.254 | 0.066 | -4.254 |
| 03 | -0.484 | -2.959 | 1.91 | -2.062 | -1.281 | -4.254 | -2.959 | -2.414 | -0.007 | -2.062 | 1.87 | -1.427 | -4.254 | -4.254 | -2.062 | -4.254 |
| 04 | -1.803 | -4.254 | 0.49 | -2.414 | -2.062 | -4.254 | -4.254 | -2.959 | -0.94 | -4.254 | 2.313 | 1.069 | -2.959 | -4.254 | -1.041 | -2.959 |
| 05 | -4.254 | -2.414 | -2.414 | -0.614 | -4.254 | -4.254 | -4.254 | -4.254 | -1.427 | -0.128 | 0.62 | 2.211 | -2.959 | -2.062 | -0.484 | 0.837 |
| 06 | -2.062 | -2.959 | -2.414 | -2.959 | -1.597 | -0.217 | -4.254 | -2.414 | -2.062 | 0.735 | -1.427 | -2.062 | -0.94 | 2.065 | 0.96 | 0.101 |
| 07 | -0.848 | -4.254 | -1.281 | -2.062 | 2.235 | -1.597 | -0.315 | -0.614 | 0.679 | -1.427 | -0.484 | -2.062 | 0.03 | -2.414 | -1.597 | -2.959 |
| 08 | 0.698 | 1.511 | 1.349 | 0.87 | -2.414 | -1.281 | -2.959 | -2.062 | -1.427 | -0.172 | -0.764 | -1.281 | -4.254 | -0.94 | -2.414 | -1.041 |
| 09 | -0.265 | -0.686 | -1.597 | -0.128 | 0.443 | 0.066 | -1.041 | 1.12 | -0.424 | 0.198 | 0.066 | 0.419 | -0.686 | -0.547 | -0.172 | 0.198 |
| 10 | -2.062 | -1.281 | 1.107 | -2.959 | 0.166 | -0.94 | 0.394 | -1.154 | -1.154 | -0.94 | 0.535 | -2.959 | -0.94 | -0.315 | 1.707 | -2.959 |
| 11 | -0.128 | -2.959 | 0.066 | -4.254 | 0.258 | -4.254 | -0.94 | -2.414 | 1.72 | -2.062 | 1.753 | -1.427 | -2.414 | -4.254 | -1.041 | -4.254 |
| 12 | -4.254 | -4.254 | 2.055 | -4.254 | -4.254 | -4.254 | -1.803 | -4.254 | -1.597 | -4.254 | 1.975 | -1.803 | -4.254 | -4.254 | -1.154 | -4.254 |
| 13 | -4.254 | -4.254 | -1.597 | -4.254 | -4.254 | -4.254 | -4.254 | -4.254 | -2.414 | -1.281 | 1.636 | 2.302 | -4.254 | -4.254 | -2.062 | -2.959 |
| 14 | -2.959 | -4.254 | -2.959 | -4.254 | -2.414 | -2.414 | -2.959 | -2.414 | -0.265 | -0.315 | 0.101 | 1.056 | -0.686 | -0.315 | 0.166 | 2.035 |
| 15 | -4.254 | 0.198 | -2.414 | -2.414 | -2.959 | -0.007 | -2.959 | -0.764 | -2.959 | 0.579 | -1.427 | -1.154 | -4.254 | 1.985 | -0.484 | 1.002 |
| 16 | -4.254 | -4.254 | -2.414 | -4.254 | 2.129 | -0.484 | 0.134 | 0.101 | -0.547 | -1.597 | -2.414 | -2.062 | -0.007 | -0.265 | 0.394 | -1.041 |
| 17 | 0.716 | 0.821 | 0.945 | 1.132 | -0.848 | -0.764 | -2.062 | -0.547 | -0.686 | -0.848 | -0.764 | 0.341 | -1.154 | -0.764 | -1.427 | -0.547 |