Model info
| Transcription factor | Batf | ||||||||
| Model | BATF_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 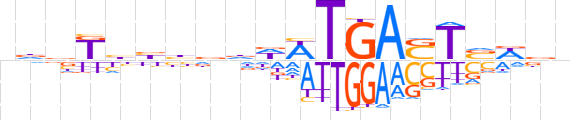 | ||||||||
| LOGO (reverse complement) | 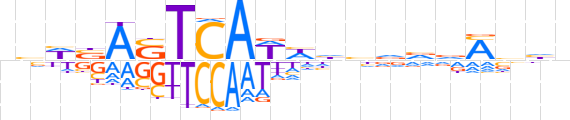 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 20 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndbKhbhddWWTGASTvWbn | ||||||||
| Best auROC (human) | 0.975 | ||||||||
| Best auROC (mouse) | 0.97 | ||||||||
| Peak sets in benchmark (human) | 10 | ||||||||
| Peak sets in benchmark (mouse) | 20 | ||||||||
| Aligned words | 507 | ||||||||
| TF family | B-ATF-related factors {1.1.4} | ||||||||
| TF subfamily | B-ATF {1.1.4.0.1} | ||||||||
| MGI | MGI:1859147 | ||||||||
| EntrezGene | GeneID:53314 (SSTAR profile) | ||||||||
| UniProt ID | BATF_MOUSE | ||||||||
| UniProt AC | O35284 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Batf expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 59.0 | 13.0 | 46.0 | 18.0 | 51.0 | 9.0 | 7.0 | 39.0 | 49.0 | 5.0 | 41.0 | 19.0 | 37.0 | 14.0 | 70.0 | 18.0 |
| 02 | 25.0 | 44.0 | 99.0 | 28.0 | 15.0 | 14.0 | 2.0 | 10.0 | 27.0 | 33.0 | 68.0 | 36.0 | 8.0 | 18.0 | 38.0 | 30.0 |
| 03 | 3.0 | 12.0 | 16.0 | 44.0 | 5.0 | 1.0 | 3.0 | 100.0 | 6.0 | 15.0 | 43.0 | 143.0 | 4.0 | 3.0 | 17.0 | 80.0 |
| 04 | 6.0 | 2.0 | 6.0 | 4.0 | 8.0 | 8.0 | 3.0 | 12.0 | 24.0 | 26.0 | 10.0 | 19.0 | 47.0 | 136.0 | 27.0 | 157.0 |
| 05 | 12.0 | 40.0 | 19.0 | 14.0 | 21.0 | 46.0 | 8.0 | 97.0 | 5.0 | 24.0 | 8.0 | 9.0 | 12.0 | 26.0 | 25.0 | 129.0 |
| 06 | 5.0 | 33.0 | 6.0 | 6.0 | 47.0 | 32.0 | 0.0 | 57.0 | 7.0 | 19.0 | 13.0 | 21.0 | 15.0 | 134.0 | 41.0 | 59.0 |
| 07 | 2.0 | 15.0 | 56.0 | 1.0 | 95.0 | 30.0 | 33.0 | 60.0 | 13.0 | 6.0 | 28.0 | 13.0 | 19.0 | 21.0 | 89.0 | 14.0 |
| 08 | 45.0 | 19.0 | 42.0 | 23.0 | 43.0 | 11.0 | 6.0 | 12.0 | 95.0 | 11.0 | 48.0 | 52.0 | 33.0 | 12.0 | 27.0 | 16.0 |
| 09 | 99.0 | 3.0 | 56.0 | 58.0 | 9.0 | 1.0 | 7.0 | 36.0 | 51.0 | 2.0 | 20.0 | 50.0 | 7.0 | 0.0 | 6.0 | 90.0 |
| 10 | 147.0 | 6.0 | 2.0 | 11.0 | 4.0 | 1.0 | 0.0 | 1.0 | 80.0 | 3.0 | 0.0 | 6.0 | 99.0 | 52.0 | 3.0 | 80.0 |
| 11 | 0.0 | 0.0 | 0.0 | 330.0 | 0.0 | 0.0 | 0.0 | 62.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 1.0 | 0.0 | 97.0 |
| 12 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.0 | 13.0 | 442.0 | 31.0 |
| 13 | 7.0 | 1.0 | 0.0 | 0.0 | 14.0 | 0.0 | 0.0 | 0.0 | 434.0 | 4.0 | 0.0 | 4.0 | 30.0 | 0.0 | 1.0 | 0.0 |
| 14 | 27.0 | 293.0 | 158.0 | 7.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 |
| 15 | 2.0 | 1.0 | 0.0 | 24.0 | 36.0 | 2.0 | 3.0 | 258.0 | 13.0 | 4.0 | 3.0 | 142.0 | 1.0 | 0.0 | 4.0 | 2.0 |
| 16 | 2.0 | 23.0 | 27.0 | 0.0 | 4.0 | 2.0 | 1.0 | 0.0 | 2.0 | 4.0 | 1.0 | 3.0 | 69.0 | 220.0 | 119.0 | 18.0 |
| 17 | 39.0 | 6.0 | 18.0 | 14.0 | 209.0 | 11.0 | 8.0 | 21.0 | 34.0 | 11.0 | 29.0 | 74.0 | 6.0 | 6.0 | 3.0 | 6.0 |
| 18 | 26.0 | 66.0 | 139.0 | 57.0 | 10.0 | 5.0 | 4.0 | 15.0 | 11.0 | 9.0 | 23.0 | 15.0 | 24.0 | 13.0 | 44.0 | 34.0 |
| 19 | 24.0 | 21.0 | 21.0 | 5.0 | 34.0 | 14.0 | 5.0 | 40.0 | 43.0 | 58.0 | 60.0 | 49.0 | 21.0 | 36.0 | 37.0 | 27.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.64 | -0.85 | 0.393 | -0.533 | 0.495 | -1.205 | -1.445 | 0.229 | 0.455 | -1.76 | 0.279 | -0.48 | 0.177 | -0.778 | 0.81 | -0.533 |
| 02 | -0.21 | 0.349 | 1.155 | -0.098 | -0.711 | -0.778 | -2.574 | -1.104 | -0.134 | 0.064 | 0.781 | 0.15 | -1.318 | -0.533 | 0.203 | -0.03 |
| 03 | -2.224 | -0.928 | -0.648 | 0.349 | -1.76 | -3.117 | -2.224 | 1.165 | -1.59 | -0.711 | 0.326 | 1.521 | -1.966 | -2.224 | -0.589 | 0.942 |
| 04 | -1.59 | -2.574 | -1.59 | -1.966 | -1.318 | -1.318 | -2.224 | -0.928 | -0.25 | -0.172 | -1.104 | -0.48 | 0.414 | 1.471 | -0.134 | 1.614 |
| 05 | -0.928 | 0.254 | -0.48 | -0.778 | -0.382 | 0.393 | -1.318 | 1.134 | -1.76 | -0.25 | -1.318 | -1.205 | -0.928 | -0.172 | -0.21 | 1.418 |
| 06 | -1.76 | 0.064 | -1.59 | -1.59 | 0.414 | 0.033 | -4.392 | 0.605 | -1.445 | -0.48 | -0.85 | -0.382 | -0.711 | 1.456 | 0.279 | 0.64 |
| 07 | -2.574 | -0.711 | 0.588 | -3.117 | 1.114 | -0.03 | 0.064 | 0.656 | -0.85 | -1.59 | -0.098 | -0.85 | -0.48 | -0.382 | 1.049 | -0.778 |
| 08 | 0.371 | -0.48 | 0.302 | -0.292 | 0.326 | -1.012 | -1.59 | -0.928 | 1.114 | -1.012 | 0.435 | 0.514 | 0.064 | -0.928 | -0.134 | -0.648 |
| 09 | 1.155 | -2.224 | 0.588 | 0.623 | -1.205 | -3.117 | -1.445 | 0.15 | 0.495 | -2.574 | -0.429 | 0.475 | -1.445 | -4.392 | -1.59 | 1.06 |
| 10 | 1.549 | -1.59 | -2.574 | -1.012 | -1.966 | -3.117 | -4.392 | -3.117 | 0.942 | -2.224 | -4.392 | -1.59 | 1.155 | 0.514 | -2.224 | 0.942 |
| 11 | -4.392 | -4.392 | -4.392 | 2.356 | -4.392 | -4.392 | -4.392 | 0.689 | -4.392 | -4.392 | -4.392 | -1.76 | -4.392 | -3.117 | -4.392 | 1.134 |
| 12 | -4.392 | -4.392 | -4.392 | -4.392 | -4.392 | -3.117 | -4.392 | -4.392 | -4.392 | -4.392 | -4.392 | -4.392 | -1.318 | -0.85 | 2.648 | 0.002 |
| 13 | -1.445 | -3.117 | -4.392 | -4.392 | -0.778 | -4.392 | -4.392 | -4.392 | 2.63 | -1.966 | -4.392 | -1.966 | -0.03 | -4.392 | -3.117 | -4.392 |
| 14 | -0.134 | 2.237 | 1.621 | -1.445 | -4.392 | -1.76 | -4.392 | -4.392 | -4.392 | -3.117 | -4.392 | -4.392 | -4.392 | -4.392 | -1.966 | -4.392 |
| 15 | -2.574 | -3.117 | -4.392 | -0.25 | 0.15 | -2.574 | -2.224 | 2.11 | -0.85 | -1.966 | -2.224 | 1.514 | -3.117 | -4.392 | -1.966 | -2.574 |
| 16 | -2.574 | -0.292 | -0.134 | -4.392 | -1.966 | -2.574 | -3.117 | -4.392 | -2.574 | -1.966 | -3.117 | -2.224 | 0.795 | 1.951 | 1.338 | -0.533 |
| 17 | 0.229 | -1.59 | -0.533 | -0.778 | 1.9 | -1.012 | -1.318 | -0.382 | 0.093 | -1.012 | -0.064 | 0.865 | -1.59 | -1.59 | -2.224 | -1.59 |
| 18 | -0.172 | 0.751 | 1.493 | 0.605 | -1.104 | -1.76 | -1.966 | -0.711 | -1.012 | -1.205 | -0.292 | -0.711 | -0.25 | -0.85 | 0.349 | 0.093 |
| 19 | -0.25 | -0.382 | -0.382 | -1.76 | 0.093 | -0.778 | -1.76 | 0.254 | 0.326 | 0.623 | 0.656 | 0.455 | -0.382 | 0.15 | 0.177 | -0.134 |