Dinucleotide PWMs for MOUSE transcription factors
| Model | LOGO | Transcription factor | Model length | Quality | Model rank | Consensus | Model release | Data source | Best auROC (human) | Best auROC (mouse) | Peak sets in benchmark (human) | Peak sets in benchmark (mouse) | Aligned words | TF family | TF subfamily | MGI | EntrezGene | UniProt ID | UniProt AC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANDR_MOUSE.H11DI.0.A | 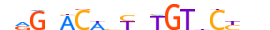 |
MOUSE:Ar | 18 | A | 0 | nRGdACWbTbTGTbCYbn | HOCOMOCOv11 | ChIP-Seq | 0.976 | 0.995 | 461 | 57 | 512 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 88064 | 11835 | ANDR_MOUSE | P19091 |
| AP2A_MOUSE.H11DI.0.A |  |
MOUSE:Tfap2a | 16 | A | 0 | nvbdSCCYGRGGCnvn | HOCOMOCOv11 | ChIP-Seq | 0.954 | 0.964 | 13 | 13 | 501 | AP-2{1.3.1} | AP-2alpha{1.3.1.0.1} | 104671 | 21418 | AP2A_MOUSE | P34056 |
| AP2C_MOUSE.H11DI.0.A |  |
MOUSE:Tfap2c | 17 | A | 0 | nhnbnSCCYCAGGShnn | HOCOMOCOv11 | ChIP-Seq | 0.954 | 0.962 | 18 | 6 | 493 | AP-2{1.3.1} | AP-2gamma{1.3.1.0.3} | 106032 | 21420 | AP2C_MOUSE | Q61312 |
| ASCL1_MOUSE.H11DI.0.A |  |
MOUSE:Ascl1 | 14 | A | 0 | nvCAGCTGCYbnbn | HOCOMOCOv11 | ChIP-Seq | 0.976 | 0.993 | 30 | 25 | 342 | MyoD / ASC-related factors{1.2.2} | Achaete-Scute-like factors{1.2.2.2} | 96919 | 17172 | ASCL1_MOUSE | Q02067 |
| ASCL2_MOUSE.H11DI.0.C |  |
MOUSE:Ascl2 | 13 | C | 0 | nvCASCTGCYnbn | HOCOMOCOv11 | ChIP-Seq | 0.624 | 0.933 | 8 | 9 | 522 | MyoD / ASC-related factors{1.2.2} | Achaete-Scute-like factors{1.2.2.2} | 96920 | 17173 | ASCL2_MOUSE | O35885 |
| ATF2_MOUSE.H11DI.0.A |  |
MOUSE:Atf2 | 13 | A | 0 | ndRTGAbSWMAbn | HOCOMOCOv11 | ChIP-Seq | 0.765 | 0.855 | 8 | 4 | 508 | Jun-related factors{1.1.1} | ATF-2-like factors{1.1.1.3} | 109349 | 102641666; 11909 | ATF2_MOUSE | P16951 |
| ATF3_MOUSE.H11DI.0.A |  |
MOUSE:Atf3 | 15 | A | 0 | ndRRdRRWGASWMAn | HOCOMOCOv11 | ChIP-Seq | 0.876 | 0.934 | 36 | 16 | 531 | Fos-related factors{1.1.2} | ATF-3-like factors{1.1.2.2} | 109384 | 11910 | ATF3_MOUSE | Q60765 |
| ATF4_MOUSE.H11DI.0.A |  |
MOUSE:Atf4 | 15 | A | 0 | ndvMTGATGMAAbhn | HOCOMOCOv11 | ChIP-Seq | 0.997 | 0.962 | 18 | 9 | 501 | ATF-4-related factors{1.1.6} | ATF-4{1.1.6.0.1} | 88096 | 11911 | ATF4_MOUSE | Q06507 |
| ATOH1_MOUSE.H11DI.0.B |  |
MOUSE:Atoh1 | 13 | B | 0 | ndnRRCAGMTGSn | HOCOMOCOv11 | ChIP-Seq | 0.788 | 0.996 | 3 | 4 | 501 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 104654 | 11921 | ATOH1_MOUSE | P48985 |
| BACH2_MOUSE.H11DI.0.A |  |
MOUSE:Bach2 | 13 | A | 0 | nhSCYKAGTCAYn | HOCOMOCOv11 | ChIP-Seq | 0.93 | 0.991 | 10 | 3 | 504 | Jun-related factors{1.1.1} | NF-E2-like factors{1.1.1.2} | 894679 | 12014 | BACH2_MOUSE | P97303 |
| BARX1_MOUSE.H11DI.0.C |  |
MOUSE:Barx1 | 12 | C | 0 | nbWAAKKRTTTh | HOCOMOCOv11 | ChIP-Seq | 0.566 | 0.832 | 2 | 2 | 462 | NK-related factors{3.1.2} | BARX{3.1.2.2} | 103124 | 12022 | BARX1_MOUSE | Q9ER42 |
| BATF3_MOUSE.H11DI.0.A |  |
MOUSE:Batf3 | 17 | A | 0 | ndSTTYYvdWWTKAbhn | HOCOMOCOv11 | ChIP-Seq | 0.993 | 5 | 500 | B-ATF-related factors{1.1.4} | B-ATF-3{1.1.4.0.3} | 1925491 | 381319 | BATF3_MOUSE | Q9D275 | ||
| BATF_MOUSE.H11DI.0.A |  |
MOUSE:Batf | 20 | A | 0 | ndbKhbhddWWTGASTvWbn | HOCOMOCOv11 | ChIP-Seq | 0.975 | 0.97 | 10 | 20 | 507 | B-ATF-related factors{1.1.4} | B-ATF{1.1.4.0.1} | 1859147 | 53314 | BATF_MOUSE | O35284 |
| BCL6_MOUSE.H11DI.0.A |  |
MOUSE:Bcl6 | 16 | A | 0 | nndMYbTCYAGKRAhn | HOCOMOCOv10 | ChIP-Seq | 0.828 | 0.892 | 27 | 11 | 504 | More than 3 adjacent zinc finger factors{2.3.3} | BCL6 factors{2.3.3.22} | 107187 | 12053 | BCL6_MOUSE | P41183 |
| BHA15_MOUSE.H11DI.0.A |  |
MOUSE:Bhlha15 | 13 | A | 0 | nvvRRCAGCTGKn | HOCOMOCOv11 | ChIP-Seq | 0.991 | 6 | 522 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 891976 | 17341 | BHA15_MOUSE | Q9QYC3 | ||
| BHE40_MOUSE.H11DI.0.A |  |
MOUSE:Bhlhe40 | 13 | A | 0 | vhKCACGTGhShn | HOCOMOCOv11 | ChIP-Seq | 0.84 | 0.971 | 10 | 5 | 339 | Hairy-related factors{1.2.4} | Hairy-like factors{1.2.4.1} | 1097714 | 20893 | BHE40_MOUSE | O35185 |
| BMAL1_MOUSE.H11DI.0.A |  |
MOUSE:Arntl | 18 | A | 0 | nnbbvbnnSCAYGTGhbn | HOCOMOCOv11 | ChIP-Seq | 0.859 | 0.883 | 18 | 15 | 420 | PAS domain factors{1.2.5} | Arnt-like factors{1.2.5.2} | 1096381 | 11865 | BMAL1_MOUSE | Q9WTL8 |
| BRAC_MOUSE.H11DI.0.A |  |
MOUSE:T | 27 | A | 0 | nWTvdYWbbddnSTGWKARdbnnnndn | HOCOMOCOv11 | ChIP-Seq | 0.807 | 0.81 | 9 | 10 | 284 | Brachyury-related factors{6.5.1} | T (Brachyury){6.5.1.0.1} | 98472 | 20997 | BRAC_MOUSE | P20293 |
| CDX2_MOUSE.H11DI.0.A |  |
MOUSE:Cdx2 | 14 | A | 0 | nnbTTTATKRShbh | HOCOMOCOv11 | ChIP-Seq | 0.927 | 0.989 | 13 | 23 | 500 | HOX-related factors{3.1.1} | CDX (Caudal type homeobox){3.1.1.9} | 88361 | 12591 | CDX2_MOUSE | P43241 |
| CEBPA_MOUSE.H11DI.0.A | 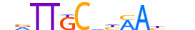 |
MOUSE:Cebpa | 13 | A | 0 | nvTTGCdhMAbhn | HOCOMOCOv11 | ChIP-Seq | 0.987 | 0.992 | 18 | 153 | 411 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 99480 | 12606 | CEBPA_MOUSE | P53566 |
| CEBPB_MOUSE.H11DI.0.A | 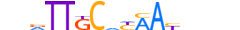 |
MOUSE:Cebpb | 16 | A | 0 | nbRTTKCdYAAYhnhn | HOCOMOCOv11 | ChIP-Seq | 0.982 | 0.989 | 65 | 158 | 505 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 88373 | 12608 | CEBPB_MOUSE | P28033 |
| CEBPG_MOUSE.H11DI.0.B |  |
MOUSE:Cebpg | 14 | B | 0 | nRTTKCATCAKhhn | HOCOMOCOv11 | ChIP-Seq | 0.702 | 0.973 | 2 | 4 | 472 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 104982 | 12611 | CEBPG_MOUSE | P53568 |
| COE1_MOUSE.H11DI.0.A |  |
MOUSE:Ebf1 | 23 | A | 0 | nvndbYCCCWKGGGRvhnnnnnn | HOCOMOCOv10 | ChIP-Seq | 0.996 | 0.994 | 17 | 55 | 506 | Early B-Cell Factor-related factors{6.1.5} | EBF1 (COE1){6.1.5.0.1} | 95275 | 13591 | COE1_MOUSE | Q07802 |
| COT2_MOUSE.H11DI.0.A |  |
MOUSE:Nr2f2 | 19 | A | 0 | nnddnnnhhvAGGTCAdvn | HOCOMOCOv10 | ChIP-Seq | 0.95 | 0.874 | 28 | 3 | 501 | RXR-related receptors (NR2){2.1.3} | COUP-like receptors (NR2F){2.1.3.5} | 1352452 | 11819 | COT2_MOUSE | P43135 |
| CREB1_MOUSE.H11DI.0.A |  |
MOUSE:Creb1 | 23 | A | 0 | nhbbbbbnbnnndRTGAYGYvRn | HOCOMOCOv10 | ChIP-Seq | 0.954 | 0.907 | 63 | 8 | 508 | CREB-related factors{1.1.7} | CREB-like factors{1.1.7.1} | 88494 | 12912 | CREB1_MOUSE | Q01147 |
| CRX_MOUSE.H11DI.0.A |  |
MOUSE:Crx | 13 | A | 0 | nvddvRGATTAdn | HOCOMOCOv11 | ChIP-Seq | 0.946 | 8 | 501 | Paired-related HD factors{3.1.3} | OTX{3.1.3.17} | 1194883 | 12951 | CRX_MOUSE | O54751 | ||
| CTCFL_MOUSE.H11DI.0.A |  |
MOUSE:Ctcfl | 21 | A | 0 | bvvCCdSYAGGKGGCGChvbn | HOCOMOCOv11 | ChIP-Seq | 0.978 | 0.997 | 25 | 8 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | CTCF-like factors{2.3.3.50} | 3652571 | 664799 | CTCFL_MOUSE | A2APF3 |
| CTCF_MOUSE.H11DI.0.A |  |
MOUSE:Ctcf | 23 | A | 0 | nbbRCCRSYAGGKGGCGSbvbnb | HOCOMOCOv11 | ChIP-Seq | 0.999 | 1.0 | 502 | 263 | 495 | More than 3 adjacent zinc finger factors{2.3.3} | CTCF-like factors{2.3.3.50} | 109447 | 13018 | CTCF_MOUSE | Q61164 |
| CUX2_MOUSE.H11DI.0.C |  |
MOUSE:Cux2 | 14 | C | 0 | ddnhAATCAATvvh | HOCOMOCOv11 | ChIP-Seq | 0.876 | 5 | 608 | HD-CUT factors{3.1.9} | CUX{3.1.9.2} | 107321 | 13048 | CUX2_MOUSE | P70298 | ||
| DDIT3_MOUSE.H11DI.0.C |  |
MOUSE:Ddit3 | 13 | C | 0 | dnvTGATGMAAbn | HOCOMOCOv11 | ChIP-Seq | 0.647 | 0.92 | 3 | 6 | 501 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 109247 | 13198 | DDIT3_MOUSE | P35639 |
| DLX5_MOUSE.H11DI.0.C |  |
MOUSE:Dlx5 | 16 | C | 0 | nWRATTRbWKbbYdbn | HOCOMOCOv11 | ChIP-Seq | 0.932 | 2 | 381 | NK-related factors{3.1.2} | DLX{3.1.2.5} | 101926 | 13395 | DLX5_MOUSE | P70396 | ||
| DMRT1_MOUSE.H11DI.0.C |  |
MOUSE:Dmrt1 | 19 | C | 0 | nhdGhhACAAWGTWdChdn | HOCOMOCOv11 | ChIP-Seq | 0.567 | 0.988 | 2 | 4 | 541 | DMRT{2.5.1} | DMRT1{2.5.1.0.1} | 1354733 | 50796 | DMRT1_MOUSE | Q9QZ59 |
| DMRTB_MOUSE.H11DI.0.C |  |
MOUSE:Dmrtb1 | 18 | C | 0 | nbdGhWACATTGTWdChn | HOCOMOCOv11 | ChIP-Seq | 0.9 | 3 | 460 | DMRT{2.5.1} | DMRTB1{2.5.1.0.6} | 1927125 | 56296 | DMRTB_MOUSE | A2A9I7 | ||
| E2F1_MOUSE.H11DI.0.A |  |
MOUSE:E2f1 | 16 | A | 0 | nddKGGCGGGRvnnvn | HOCOMOCOv11 | ChIP-Seq | 0.902 | 0.982 | 80 | 21 | 500 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 101941 | 13555 | E2F1_MOUSE | Q61501 |
| E2F3_MOUSE.H11DI.0.A |  |
MOUSE:E2f3 | 17 | A | 0 | vvvSGCGGGARvvvvvv | HOCOMOCOv11 | ChIP-Seq | 0.859 | 0.993 | 3 | 22 | 501 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 1096340 | 13557 | E2F3_MOUSE | O35261 |
| E2F4_MOUSE.H11DI.0.A |  |
MOUSE:E2f4 | 17 | A | 0 | nSGCGSSvvnKbbvvdn | HOCOMOCOv11 | ChIP-Seq | 0.957 | 0.989 | 15 | 37 | 487 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 103012 | 104394 | E2F4_MOUSE | Q8R0K9 |
| EGR1_MOUSE.H11DI.0.A |  |
MOUSE:Egr1 | 21 | A | 0 | nvbGYGKGGGYGGRRvbvbvv | HOCOMOCOv11 | ChIP-Seq | 0.991 | 0.986 | 36 | 12 | 526 | Three-zinc finger Krüppel-related factors{2.3.1} | EGR factors{2.3.1.3} | 95295 | 13653 | EGR1_MOUSE | P08046 |
| EGR2_MOUSE.H11DI.0.A |  |
MOUSE:Egr2 | 21 | A | 0 | vdvdRhGKGGGhGKvddnndv | HOCOMOCOv11 | ChIP-Seq | 0.961 | 0.996 | 4 | 25 | 575 | Three-zinc finger Krüppel-related factors{2.3.1} | EGR factors{2.3.1.3} | 95296 | 13654 | EGR2_MOUSE | P08152 |
| EHF_MOUSE.H11DI.0.C |  |
MOUSE:Ehf | 14 | C | 0 | hAbSAGGAARYdvn | HOCOMOCOv11 | ChIP-Seq | 0.964 | 0.634 | 5 | 3 | 457 | Ets-related factors{3.5.2} | EHF-like factors{3.5.2.4} | 1270840 | 13661 | EHF_MOUSE | O70273 |
| ELF1_MOUSE.H11DI.0.A |  |
MOUSE:Elf1 | 14 | A | 0 | vvdnSCGGAAGYvv | HOCOMOCOv11 | ChIP-Seq | 0.993 | 0.979 | 43 | 5 | 502 | Ets-related factors{3.5.2} | Elf-1-like factors{3.5.2.3} | 107180 | 13709 | ELF1_MOUSE | Q60775 |
| ELF5_MOUSE.H11DI.0.A |  |
MOUSE:Elf5 | 20 | A | 0 | vdvvRRRRAvvAGGAARddv | HOCOMOCOv11 | ChIP-Seq | 0.859 | 0.993 | 3 | 12 | 522 | Ets-related factors{3.5.2} | EHF-like factors{3.5.2.4} | 1335079 | 13711 | ELF5_MOUSE | Q8VDK3 |
| ELK1_MOUSE.H11DI.0.B |  |
MOUSE:Elk1 | 12 | B | 0 | nvSCGGAAGYvv | HOCOMOCOv11 | ChIP-Seq | 0.985 | 0.706 | 4 | 7 | 511 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 101833 | 13712 | ELK1_MOUSE | P41969 |
| ELK4_MOUSE.H11DI.0.B |  |
MOUSE:Elk4 | 16 | B | 0 | vvMCGGAARYvvvnnb | HOCOMOCOv11 | ChIP-Seq | 0.966 | 0.632 | 13 | 4 | 190 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 102853 | 13714 | ELK4_MOUSE | P41158 |
| ERG_MOUSE.H11DI.0.A |  |
MOUSE:Erg | 23 | A | 0 | vvvvbvvvMMGGAARbSvvbbnn | HOCOMOCOv10 | ChIP-Seq | 0.958 | 0.977 | 79 | 16 | 497 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 95415 | 13876 | ERG_MOUSE | P81270 |
| ERR1_MOUSE.H11DI.0.A |  |
MOUSE:Esrra | 16 | A | 0 | nhdbYSAAGGTCAnnn | HOCOMOCOv11 | ChIP-Seq | 0.951 | 0.989 | 31 | 3 | 533 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 1346831 | 26379 | ERR1_MOUSE | O08580 |
| ERR2_MOUSE.H11DI.0.A |  |
MOUSE:Esrrb | 23 | A | 0 | nndbnbSAAGGTCAnvbnvnvnn | HOCOMOCOv10 | ChIP-Seq | 0.994 | 14 | 529 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 1346832 | 26380 | ERR2_MOUSE | Q61539 | ||
| ERR3_MOUSE.H11DI.0.C |  |
MOUSE:Esrrg | 12 | C | 0 | nbCAAGGWSAbn | HOCOMOCOv11 | ChIP-Seq | 0.86 | 2 | 258 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 1347056 | 26381 | ERR3_MOUSE | P62509 | ||
| ESR1_MOUSE.H11DI.0.A |  |
MOUSE:Esr1 | 17 | A | 0 | nRGGKCASCvTGMCMYn | HOCOMOCOv11 | ChIP-Seq | 0.976 | 0.996 | 432 | 58 | 527 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 1352467 | 13982 | ESR1_MOUSE | P19785 |
| ESR2_MOUSE.H11DI.0.A |  |
MOUSE:Esr2 | 20 | A | 0 | nbbnRGGYCRShvTGhCCYn | HOCOMOCOv11 | ChIP-Seq | 0.926 | 0.943 | 11 | 7 | 223 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 109392 | 13983 | ESR2_MOUSE | O08537 |
| ETS1_MOUSE.H11DI.0.A |  |
MOUSE:Ets1 | 15 | A | 0 | vvvRSMGGAAGYRvn | HOCOMOCOv11 | ChIP-Seq | 0.959 | 0.966 | 45 | 33 | 511 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 95455 | 23871 | ETS1_MOUSE | P27577 |
| ETV2_MOUSE.H11DI.0.A |  |
MOUSE:Etv2 | 17 | A | 0 | vvdvRCAGGAARYdSvn | HOCOMOCOv11 | ChIP-Seq | 0.985 | 6 | 526 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 99253 | 14008 | ETV2_MOUSE | P41163 | ||
| ETV6_MOUSE.H11DI.0.C |  |
MOUSE:Etv6 | 14 | C | 0 | ndnvdvWGSAAdbn | HOCOMOCOv11 | ChIP-Seq | 0.857 | 3 | 506 | Ets-related factors{3.5.2} | ETV6-like factors{3.5.2.6} | 109336 | 14011 | ETV6_MOUSE | P97360 | ||
| FLI1_MOUSE.H11DI.0.A |  |
MOUSE:Fli1 | 16 | A | 0 | vvvSMGGAARbSvvbv | HOCOMOCOv11 | ChIP-Seq | 0.958 | 0.969 | 50 | 32 | 499 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 95554 | 14247 | FLI1_MOUSE | P26323 |
| FOSB_MOUSE.H11DI.0.A |  |
MOUSE:Fosb | 11 | A | 0 | nvTGAGTCAbn | HOCOMOCOv11 | ChIP-Seq | 0.97 | 0.919 | 2 | 27 | 523 | Fos-related factors{1.1.2} | Fos factors{1.1.2.1} | 95575 | 14282 | FOSB_MOUSE | P13346 |
| FOSL1_MOUSE.H11DI.0.A |  |
MOUSE:Fosl1 | 13 | A | 0 | bnRTGAGTCAYvn | HOCOMOCOv11 | ChIP-Seq | 0.987 | 0.953 | 21 | 2 | 175 | Fos-related factors{1.1.2} | Fos factors{1.1.2.1} | 107179 | 14283 | FOSL1_MOUSE | P48755 |
| FOSL2_MOUSE.H11DI.0.A |  |
MOUSE:Fosl2 | 15 | A | 0 | nnnbnvTGAGTCAYn | HOCOMOCOv11 | ChIP-Seq | 0.995 | 0.975 | 27 | 5 | 131 | Fos-related factors{1.1.2} | Fos factors{1.1.2.1} | 102858 | 14284 | FOSL2_MOUSE | P47930 |
| FOS_MOUSE.H11DI.0.A | 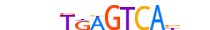 |
MOUSE:Fos | 14 | A | 0 | nbnnTRAGTCAbhn | HOCOMOCOv11 | ChIP-Seq | 0.993 | 0.929 | 38 | 50 | 500 | Fos-related factors{1.1.2} | Fos factors{1.1.2.1} | 95574 | 14281 | FOS_MOUSE | P01101 |
| FOXA1_MOUSE.H11DI.0.A |  |
MOUSE:Foxa1 | 16 | A | 0 | nbnnTGTTTACWYWdn | HOCOMOCOv11 | ChIP-Seq | 0.987 | 0.984 | 199 | 28 | 510 | Forkhead box (FOX) factors{3.3.1} | FOXA{3.3.1.1} | 1347472 | 15375 | FOXA1_MOUSE | P35582 |
| FOXA2_MOUSE.H11DI.0.A |  |
MOUSE:Foxa2 | 16 | A | 0 | nbnhTGTTTACWYWdn | HOCOMOCOv11 | ChIP-Seq | 0.985 | 0.994 | 22 | 34 | 501 | Forkhead box (FOX) factors{3.3.1} | FOXA{3.3.1.1} | 1347476 | 15376 | FOXA2_MOUSE | P35583 |
| FOXA3_MOUSE.H11DI.0.A |  |
MOUSE:Foxa3 | 16 | A | 0 | nbbhTGTTTACWYWdn | HOCOMOCOv11 | ChIP-Seq | 0.993 | 6 | 515 | Forkhead box (FOX) factors{3.3.1} | FOXA{3.3.1.1} | 1347477 | 15377 | FOXA3_MOUSE | P35584 | ||
| FOXD3_MOUSE.H11DI.0.B |  |
MOUSE:Foxd3 | 16 | B | 0 | nbnWGTTThYhYdvbn | HOCOMOCOv11 | ChIP-Seq | 0.814 | 11 | 415 | Forkhead box (FOX) factors{3.3.1} | FOXD{3.3.1.4} | 1347473 | 15221 | FOXD3_MOUSE | Q61060 | ||
| FOXK1_MOUSE.H11DI.0.A |  |
MOUSE:Foxk1 | 12 | A | 0 | nbTGTTThYnbn | HOCOMOCOv11 | ChIP-Seq | 0.86 | 0.914 | 11 | 4 | 502 | Forkhead box (FOX) factors{3.3.1} | FOXK{3.3.1.11} | 1347488 | 17425 | FOXK1_MOUSE | P42128 |
| FOXL2_MOUSE.H11DI.0.C |  |
MOUSE:Foxl2 | 14 | C | 0 | nbnTGTTTWYhhWn | HOCOMOCOv11 | ChIP-Seq | 0.943 | 2 | 500 | Forkhead box (FOX) factors{3.3.1} | FOXL{3.3.1.12} | 1349428 | 26927 | FOXL2_MOUSE | O88470 | ||
| FOXO1_MOUSE.H11DI.0.A |  |
MOUSE:Foxo1 | 11 | A | 0 | nbbTGTTTMCn | HOCOMOCOv11 | ChIP-Seq | 0.878 | 0.934 | 9 | 21 | 500 | Forkhead box (FOX) factors{3.3.1} | FOXO{3.3.1.15} | 1890077 | 56458 | FOXO1_MOUSE | Q9R1E0 |
| FOXO3_MOUSE.H11DI.0.B |  |
MOUSE:Foxo3 | 12 | B | 0 | nbbTGTTTAChn | HOCOMOCOv11 | ChIP-Seq | 0.666 | 0.953 | 2 | 7 | 500 | Forkhead box (FOX) factors{3.3.1} | FOXO{3.3.1.15} | 1890081 | 56484 | FOXO3_MOUSE | Q9WVH4 |
| GABPA_MOUSE.H11DI.0.A |  |
MOUSE:Gabpa | 15 | A | 0 | vvvSMGGAAGYRvnv | HOCOMOCOv11 | ChIP-Seq | 0.988 | 0.957 | 92 | 8 | 503 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 95610 | 14390 | GABPA_MOUSE | Q00422 |
| GATA1_MOUSE.H11DI.0.A |  |
MOUSE:Gata1 | 20 | A | 0 | nbbbnnnnndvWGATAAvvn | HOCOMOCOv11 | ChIP-Seq | 0.983 | 0.971 | 48 | 122 | 505 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 95661 | 14460 | GATA1_MOUSE | P17679 |
| GATA2_MOUSE.H11DI.0.A |  |
MOUSE:Gata2 | 22 | A | 0 | nvnnndnnnnnvWGATARvvdn | HOCOMOCOv11 | ChIP-Seq | 0.966 | 0.966 | 84 | 54 | 501 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 95662 | 14461 | GATA2_MOUSE | O09100 |
| GATA3_MOUSE.H11DI.0.A |  |
MOUSE:Gata3 | 23 | A | 0 | ndnddndbdnnvAGATAAvddnn | HOCOMOCOv10 | ChIP-Seq | 0.95 | 0.947 | 98 | 60 | 504 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 95663 | 14462 | GATA3_MOUSE | P23772 |
| GATA4_MOUSE.H11DI.0.A |  |
MOUSE:Gata4 | 15 | A | 0 | ndnndvWGATAARvn | HOCOMOCOv11 | ChIP-Seq | 0.947 | 0.949 | 15 | 17 | 428 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 95664 | 14463 | GATA4_MOUSE | Q08369 |
| GATA6_MOUSE.H11DI.0.A |  |
MOUSE:Gata6 | 10 | A | 0 | vWGATAASvn | HOCOMOCOv11 | ChIP-Seq | 0.963 | 0.97 | 29 | 7 | 503 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 107516 | 14465 | GATA6_MOUSE | Q61169 |
| GCR_MOUSE.H11DI.0.A |  |
MOUSE:Nr3c1 | 21 | A | 0 | nvvnvRGvACAvRvWGTnMhn | HOCOMOCOv11 | ChIP-Seq | 0.983 | 0.988 | 143 | 119 | 747 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 95824 | GCR_MOUSE | P06537 | |
| GFI1B_MOUSE.H11DI.0.A |  |
MOUSE:Gfi1b | 12 | A | 0 | dKCWSWGATTKn | HOCOMOCOv11 | ChIP-Seq | 0.881 | 0.899 | 3 | 24 | 524 | More than 3 adjacent zinc finger factors{2.3.3} | GFI1 factors{2.3.3.21} | 1276578 | 14582 | GFI1B_MOUSE | O70237 |
| GLI1_MOUSE.H11DI.0.B |  |
MOUSE:Gli1 | 17 | B | 0 | nvnnbSTGGGTGGYChn | HOCOMOCOv11 | ChIP-Seq | 0.671 | 0.953 | 2 | 3 | 501 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 95727 | 14632 | GLI1_MOUSE | P47806 |
| GRHL2_MOUSE.H11DI.0.A |  |
MOUSE:Grhl2 | 15 | A | 0 | RCYdGTYYddChRGh | HOCOMOCOv11 | ChIP-Seq | 0.965 | 0.982 | 22 | 4 | 501 | Grainyhead-related factors{6.7.1} | GRH-like proteins{6.7.1.1} | 2182543 | 252973 | GRHL2_MOUSE | Q8K5C0 |
| HAND1_MOUSE.H11DI.0.C | 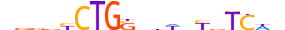 |
MOUSE:Hand1 | 19 | C | 0 | nddvYCTGShhWnYvTCWn | HOCOMOCOv11 | ChIP-Seq | 0.998 | 3 | 505 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 103577 | 15110 | HAND1_MOUSE | Q64279 | ||
| HIF1A_MOUSE.H11DI.0.C |  |
MOUSE:Hif1a | 11 | C | 0 | bnvKRCGTGhb | HOCOMOCOv11 | ChIP-Seq | 0.719 | 0.718 | 39 | 8 | 515 | PAS domain factors{1.2.5} | Ahr-like factors{1.2.5.1} | 106918 | 15251 | HIF1A_MOUSE | Q61221 |
| HLF_MOUSE.H11DI.0.C |  |
MOUSE:Hlf | 12 | C | 0 | nvTTRCAYAAbn | HOCOMOCOv11 | ChIP-Seq | 0.973 | 4 | 514 | C/EBP-related{1.1.8} | PAR factors{1.1.8.2} | 96108 | 217082 | HLF_MOUSE | Q8BW74 | ||
| HNF1B_MOUSE.H11DI.0.A |  |
MOUSE:Hnf1b | 17 | A | 0 | ndRTWAATGWTTRMYhn | HOCOMOCOv11 | ChIP-Seq | 0.953 | 0.997 | 5 | 8 | 500 | POU domain factors{3.1.10} | HNF1-like factors{3.1.10.7} | 98505 | 21410 | HNF1B_MOUSE | P27889 |
| HNF4A_MOUSE.H11DI.0.A |  |
MOUSE:Hnf4a | 17 | A | 0 | hhvRRdbCARAGKbCWn | HOCOMOCOv11 | ChIP-Seq | 0.987 | 0.988 | 49 | 33 | 386 | RXR-related receptors (NR2){2.1.3} | HNF-4 (NR2A){2.1.3.2} | 109128 | 15378 | HNF4A_MOUSE | P49698 |
| HNF6_MOUSE.H11DI.0.A |  |
MOUSE:Onecut1 | 18 | A | 0 | dbTATTGATTddYbdddn | HOCOMOCOv11 | ChIP-Seq | 0.977 | 24 | 306 | HD-CUT factors{3.1.9} | ONECUT{3.1.9.1} | 1196423 | 15379 | HNF6_MOUSE | O08755 | ||
| HSF1_MOUSE.H11DI.0.A |  |
MOUSE:Hsf1 | 18 | A | 0 | nvhRGAAnnWTCYRGAAn | HOCOMOCOv11 | ChIP-Seq | 0.996 | 0.992 | 56 | 32 | 505 | HSF factors{3.4.1} | HSF1 (HSTF1){3.4.1.0.1} | 96238 | 15499 | HSF1_MOUSE | P38532 |
| HTF4_MOUSE.H11DI.0.A |  |
MOUSE:Tcf12 | 15 | A | 0 | nvCAGSTGKKnnndn | HOCOMOCOv11 | ChIP-Seq | 0.918 | 0.988 | 49 | 10 | 529 | E2A-related factors{1.2.1} | HTF-4 (TCF-12, HEB){1.2.1.0.3} | 101877 | 21406 | HTF4_MOUSE | Q61286 |
| HXA9_MOUSE.H11DI.0.A |  |
MOUSE:Hoxa9 | 14 | A | 0 | ndTGAYKKATKdbn | HOCOMOCOv11 | ChIP-Seq | 0.906 | 0.819 | 5 | 8 | 503 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 96180 | 15405 | HXA9_MOUSE | P09631 |
| HXB4_MOUSE.H11DI.0.A |  |
MOUSE:Hoxb4 | 11 | A | 0 | WKRYWRATKRS | HOCOMOCOv11 | ChIP-Seq | 0.503 | 0.921 | 2 | 12 | 503 | HOX-related factors{3.1.1} | HOX4{3.1.1.4} | 96185 | 15412 | HXB4_MOUSE | P10284 |
| HXC9_MOUSE.H11DI.0.A |  |
MOUSE:Hoxc9 | 14 | A | 0 | nWRATTTAYdRbbn | HOCOMOCOv11 | ChIP-Seq | 0.958 | 7 | 500 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 96199 | 15427 | HXC9_MOUSE | P09633 | ||
| IRF1_MOUSE.H11DI.0.A | 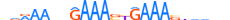 |
MOUSE:Irf1 | 23 | A | 0 | nnRAAnhGAAASYGAAAShvdvn | HOCOMOCOv11 | ChIP-Seq | 0.997 | 0.998 | 23 | 18 | 453 | Interferon-regulatory factors{3.5.3} | IRF-1{3.5.3.0.1} | 96590 | 16362 | IRF1_MOUSE | P15314 |
| IRF3_MOUSE.H11DI.0.A |  |
MOUSE:Irf3 | 23 | A | 0 | nvdRRMdRGAAAvnGAAAvndvv | HOCOMOCOv11 | ChIP-Seq | 0.797 | 0.962 | 6 | 21 | 510 | Interferon-regulatory factors{3.5.3} | IRF-3{3.5.3.0.3} | 1859179 | 54131 | IRF3_MOUSE | P70671 |
| IRF4_MOUSE.H11DI.0.A |  |
MOUSE:Irf4 | 20 | A | 0 | nddWWvvRGAASTGARAvhn | HOCOMOCOv11 | ChIP-Seq | 0.948 | 0.981 | 19 | 61 | 502 | Interferon-regulatory factors{3.5.3} | IRF-4 (LSIRF, NF-EM5, MUM1, Pip){3.5.3.0.4} | 1096873 | 16364 | IRF4_MOUSE | Q64287 |
| IRF8_MOUSE.H11DI.0.A | 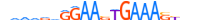 |
MOUSE:Irf8 | 22 | A | 0 | ddWWvdGGAASTGAAASYvvvd | HOCOMOCOv11 | ChIP-Seq | 0.997 | 59 | 513 | Interferon-regulatory factors{3.5.3} | IRF-8 (ICSBP1){3.5.3.0.8} | 96395 | 15900 | IRF8_MOUSE | P23611 | ||
| ISL1_MOUSE.H11DI.0.A |  |
MOUSE:Isl1 | 14 | A | 0 | nnYTAAKKRvhYhn | HOCOMOCOv11 | ChIP-Seq | 0.838 | 0.918 | 3 | 13 | 472 | HD-LIM factors{3.1.5} | ISL{3.1.5.1} | 101791 | 16392 | ISL1_MOUSE | P61372 |
| JUNB_MOUSE.H11DI.0.A |  |
MOUSE:Junb | 10 | A | 0 | vTGAGTCAbn | HOCOMOCOv11 | ChIP-Seq | 0.976 | 0.937 | 8 | 25 | 518 | Jun-related factors{1.1.1} | Jun factors{1.1.1.1} | 96647 | 16477 | JUNB_MOUSE | P09450 |
| JUND_MOUSE.H11DI.0.A |  |
MOUSE:Jund | 13 | A | 0 | nvdvTGAGTCAYn | HOCOMOCOv11 | ChIP-Seq | 0.996 | 0.988 | 64 | 21 | 400 | Jun-related factors{1.1.1} | Jun factors{1.1.1.1} | 96648 | 16478 | JUND_MOUSE | P15066 |
| JUN_MOUSE.H11DI.0.A |  |
MOUSE:Jun | 13 | A | 0 | nbnRTGAGTCAYn | HOCOMOCOv11 | ChIP-Seq | 0.944 | 0.94 | 56 | 39 | 481 | Jun-related factors{1.1.1} | Jun factors{1.1.1.1} | 96646 | 16476 | JUN_MOUSE | P05627 |
| KLF15_MOUSE.H11DI.0.A |  |
MOUSE:Klf15 | 22 | A | 0 | dRdRGGMGGRGvdRGRRRdSRv | HOCOMOCOv11 | ChIP-Seq | 0.967 | 0.998 | 4 | 3 | 530 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1929988 | 66277 | KLF15_MOUSE | Q9EPW2 |
| KLF1_MOUSE.H11DI.0.A |  |
MOUSE:Klf1 | 19 | A | 0 | ndGGGYGKGGMnKSKnvbb | HOCOMOCOv11 | ChIP-Seq | 0.915 | 0.988 | 5 | 6 | 585 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1342771 | KLF1_MOUSE | P46099 | |
| KLF3_MOUSE.H11DI.0.A |  |
MOUSE:Klf3 | 17 | A | 0 | nvnvndGGGYGKGGYhn | HOCOMOCOv11 | ChIP-Seq | 0.783 | 0.996 | 2 | 7 | 501 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1342773 | 16599 | KLF3_MOUSE | Q60980 |
| KLF4_MOUSE.H11DI.0.A |  |
MOUSE:Klf4 | 20 | A | 0 | vdvdvdGGGYGKGGbnbvbv | HOCOMOCOv11 | ChIP-Seq | 0.889 | 0.986 | 11 | 34 | 501 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1342287 | 16600 | KLF4_MOUSE | Q60793 |
| KLF5_MOUSE.H11DI.0.A |  |
MOUSE:Klf5 | 18 | A | 0 | vvvvdGGGYGGGGMndSv | HOCOMOCOv11 | ChIP-Seq | 0.96 | 0.988 | 32 | 23 | 504 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1338056 | 12224 | KLF5_MOUSE | Q9Z0Z7 |
| KLF6_MOUSE.H11DI.0.B |  |
MOUSE:Klf6 | 21 | B | 0 | vbvvndGGGYGKRRbbvnvvb | HOCOMOCOv11 | ChIP-Seq | 0.975 | 0.684 | 16 | 2 | 497 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1346318 | KLF6_MOUSE | O08584 | |
| LHX2_MOUSE.H11DI.0.A |  |
MOUSE:Lhx2 | 11 | A | 0 | nhTAATKAGvn | HOCOMOCOv11 | ChIP-Seq | 0.804 | 0.934 | 5 | 12 | 531 | HD-LIM factors{3.1.5} | Lhx-2-like factors{3.1.5.3} | 96785 | 16870 | LHX2_MOUSE | Q9Z0S2 |
| LHX3_MOUSE.H11DI.0.C |  |
MOUSE:Lhx3 | 14 | C | 0 | hhdhnbTAATKRvd | HOCOMOCOv11 | ChIP-Seq | 0.951 | 2 | 387 | HD-LIM factors{3.1.5} | Lhx-3-like factors{3.1.5.4} | 102673 | 16871 | LHX3_MOUSE | P50481 | ||
| LHX6_MOUSE.H11DI.0.C |  |
MOUSE:Lhx6 | 11 | C | 0 | nbYTGATTRvn | HOCOMOCOv11 | ChIP-Seq | 0.337 | 0.932 | 1 | 2 | 445 | HD-LIM factors{3.1.5} | Lhx-6-like factors{3.1.5.5} | 1306803 | 16874 | LHX6_MOUSE | Q9R1R0 |
| LYL1_MOUSE.H11DI.0.A |  |
MOUSE:Lyl1 | 13 | A | 0 | nhYWbCTSbbnhn | HOCOMOCOv11 | ChIP-Seq | 0.884 | 0.854 | 23 | 3 | 476 | Tal-related factors{1.2.3} | Tal / HEN-like factors{1.2.3.1} | 96891 | 17095 | LYL1_MOUSE | P27792 |
| MAFB_MOUSE.H11DI.0.B |  |
MOUSE:Mafb | 16 | B | 0 | dhnWGCTGAShhddbn | HOCOMOCOv11 | ChIP-Seq | 0.68 | 0.969 | 5 | 3 | 502 | Maf-related factors{1.1.3} | Large Maf factors{1.1.3.1} | 104555 | 16658 | MAFB_MOUSE | P54841 |
| MAFF_MOUSE.H11DI.0.A |  |
MOUSE:Maff | 23 | A | 0 | ndddnWGMTGACThWGCMnhhhn | HOCOMOCOv11 | ChIP-Seq | 0.462 | 0.983 | 2 | 15 | 500 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 96910 | 17133 | MAFF_MOUSE | O54791 |
| MAFG_MOUSE.H11DI.0.A |  |
MOUSE:Mafg | 18 | A | 0 | WGMYRASTCAGCAnWWhh | HOCOMOCOv11 | ChIP-Seq | 0.899 | 1.0 | 3 | 6 | 456 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 96911 | 17134 | MAFG_MOUSE | O54790 |
| MAFK_MOUSE.H11DI.0.A |  |
MOUSE:Mafk | 18 | A | 0 | ndWWnTGCTGASTCAKCW | HOCOMOCOv11 | ChIP-Seq | 0.996 | 0.996 | 12 | 11 | 415 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 99951 | 17135 | MAFK_MOUSE | Q61827 |
| MAF_MOUSE.H11DI.0.A |  |
MOUSE:Maf | 23 | A | 0 | ndddnTGCTGAbdhhdbhvndhn | HOCOMOCOv11 | ChIP-Seq | 0.899 | 0.909 | 10 | 5 | 454 | Maf-related factors{1.1.3} | Large Maf factors{1.1.3.1} | 96909 | 17132 | MAF_MOUSE | P54843 |
| MAX_MOUSE.H11DI.0.A |  |
MOUSE:Max | 23 | A | 0 | nvnnnnnvnvnvdvCACGTGbbn | HOCOMOCOv10 | ChIP-Seq | 0.955 | 0.968 | 86 | 14 | 514 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 96921 | MAX_MOUSE | P28574 | |
| MAZ_MOUSE.H11DI.0.A |  |
MOUSE:Maz | 25 | A | 0 | ndRGRGGWGGRRRnRvRdRvdvvvv | HOCOMOCOv11 | ChIP-Seq | 0.944 | 0.967 | 3 | 5 | 522 | Factors with multiple dispersed zinc fingers{2.3.4} | MAZ-like factors{2.3.4.8} | 1338823 | MAZ_MOUSE | P56671 | |
| MEF2A_MOUSE.H11DI.0.A |  |
MOUSE:Mef2a | 15 | A | 0 | nddYTATTTWWRRhn | HOCOMOCOv11 | ChIP-Seq | 0.948 | 0.991 | 15 | 6 | 487 | Regulators of differentiation{5.1.1} | MEF-2{5.1.1.1} | 99532 | 17258 | MEF2A_MOUSE | Q60929 |
| MEF2C_MOUSE.H11DI.0.A |  |
MOUSE:Mef2c | 14 | A | 0 | ndhTATTTWWRdhh | HOCOMOCOv11 | ChIP-Seq | 0.949 | 0.934 | 6 | 9 | 499 | Regulators of differentiation{5.1.1} | MEF-2{5.1.1.1} | 99458 | 17260 | MEF2C_MOUSE | Q8CFN5 |
| MEF2D_MOUSE.H11DI.0.A |  |
MOUSE:Mef2d | 15 | A | 0 | nndKCTATTTWTAKv | HOCOMOCOv11 | ChIP-Seq | 0.808 | 0.973 | 2 | 27 | 373 | Regulators of differentiation{5.1.1} | MEF-2{5.1.1.1} | 99533 | 17261 | MEF2D_MOUSE | Q63943 |
| MEIS1_MOUSE.H11DI.0.A |  |
MOUSE:Meis1 | 14 | A | 0 | ndTGAYTdATKdbn | HOCOMOCOv11 | ChIP-Seq | 0.848 | 0.952 | 2 | 37 | 500 | TALE-type homeo domain factors{3.1.4} | MEIS{3.1.4.2} | 104717 | 17268 | MEIS1_MOUSE | Q60954 |
| MITF_MOUSE.H11DI.0.A |  |
MOUSE:Mitf | 12 | A | 0 | nbChYGTGAShn | HOCOMOCOv11 | ChIP-Seq | 0.995 | 0.839 | 28 | 3 | 507 | bHLH-ZIP factors{1.2.6} | TFE3-like factors{1.2.6.1} | 104554 | 17342 | MITF_MOUSE | Q08874 |
| MSGN1_MOUSE.H11DI.0.C |  |
MOUSE:Msgn1 | 14 | C | 0 | nvCCATTTGKYhhn | HOCOMOCOv11 | ChIP-Seq | 0.999 | 4 | 500 | Tal-related factors{1.2.3} | Mesp-like factors{1.2.3.3} | 1860483 | 56184 | MSGN1_MOUSE | Q9JK54 | ||
| MXI1_MOUSE.H11DI.0.A |  |
MOUSE:Mxi1 | 23 | A | 0 | nvvvCAYGTGSvnvvnbvvvnbn | HOCOMOCOv10 | ChIP-Seq | 0.897 | 0.981 | 10 | 5 | 500 | bHLH-ZIP factors{1.2.6} | Mad-like factors{1.2.6.7} | 97245 | 17859 | MXI1_MOUSE | P50540 |
| MYBA_MOUSE.H11DI.0.C |  |
MOUSE:Mybl1 | 12 | C | 0 | bnRRCvGTTRRn | HOCOMOCOv11 | ChIP-Seq | 0.415 | 0.931 | 3 | 2 | 393 | Myb/SANT domain factors{3.5.1} | Myb-like factors{3.5.1.1} | 99925 | 17864 | MYBA_MOUSE | P51960 |
| MYB_MOUSE.H11DI.0.A |  |
MOUSE:Myb | 12 | A | 0 | nbddChGTTdvn | HOCOMOCOv11 | ChIP-Seq | 0.811 | 0.935 | 27 | 9 | 465 | Myb/SANT domain factors{3.5.1} | Myb-like factors{3.5.1.1} | 97249 | 17863 | MYB_MOUSE | P06876 |
| MYCN_MOUSE.H11DI.0.A |  |
MOUSE:Mycn | 15 | A | 0 | vRvbMACRWGSnvvv | HOCOMOCOv11 | ChIP-Seq | 0.921 | 0.961 | 32 | 10 | 493 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 97357 | 18109 | MYCN_MOUSE | P03966 |
| MYC_MOUSE.H11DI.0.A |  |
MOUSE:Myc | 15 | A | 0 | bvvvvvvMCACGTGb | HOCOMOCOv11 | ChIP-Seq | 0.909 | 0.963 | 265 | 126 | 450 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 97250 | 17869 | MYC_MOUSE | P01108 |
| MYOD1_MOUSE.H11DI.0.A |  |
MOUSE:Myod1 | 23 | A | 0 | nnnvCAGCTGYYbbhbbbnnbbn | HOCOMOCOv10 | ChIP-Seq | 0.993 | 0.998 | 9 | 63 | 500 | MyoD / ASC-related factors{1.2.2} | Myogenic transcription factors{1.2.2.1} | 97275 | 17927 | MYOD1_MOUSE | P10085 |
| MYOG_MOUSE.H11DI.0.A |  |
MOUSE:Myog | 22 | A | 0 | nvvvvnRRCAGCTGbndnnnnn | HOCOMOCOv10 | ChIP-Seq | 0.997 | 20 | 461 | MyoD / ASC-related factors{1.2.2} | Myogenic transcription factors{1.2.2.1} | 97276 | 17928 | MYOG_MOUSE | P12979 | ||
| NANOG_MOUSE.H11DI.0.A |  |
MOUSE:Nanog | 22 | A | 0 | nbbYWTTvWnWTKYWRAdndnn | HOCOMOCOv10 | ChIP-Seq | 0.924 | 0.944 | 15 | 38 | 513 | NK-related factors{3.1.2} | NANOG{3.1.2.12} | 1919200 | 71950 | NANOG_MOUSE | Q80Z64 |
| NDF1_MOUSE.H11DI.0.A | 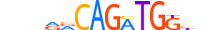 |
MOUSE:Neurod1 | 14 | A | 0 | nvnRRCAGATGGbn | HOCOMOCOv11 | ChIP-Seq | 0.949 | 0.96 | 5 | 2 | 507 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 1339708 | 18012 | NDF1_MOUSE | Q60867 |
| NDF2_MOUSE.H11DI.0.A |  |
MOUSE:Neurod2 | 15 | A | 0 | dvvvnRRCAGATGGn | HOCOMOCOv11 | ChIP-Seq | 0.619 | 0.97 | 2 | 21 | 560 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 107755 | 18013 | NDF2_MOUSE | Q62414 |
| NF2L2_MOUSE.H11DI.0.A | 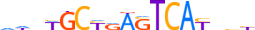 |
MOUSE:Nfe2l2 | 17 | A | 0 | dnbWGCYRASTCAYbbb | HOCOMOCOv11 | ChIP-Seq | 0.901 | 0.99 | 5 | 26 | 104 | Jun-related factors{1.1.1} | NF-E2-like factors{1.1.1.2} | 108420 | 18024 | NF2L2_MOUSE | Q60795 |
| NFAC1_MOUSE.H11DI.0.A |  |
MOUSE:Nfatc1 | 18 | A | 0 | vWGGAARvdnWKWvWSRn | HOCOMOCOv11 | ChIP-Seq | 0.726 | 0.963 | 7 | 8 | 508 | NFAT-related factors{6.1.3} | NFATc1{6.1.3.0.1} | 102469 | NFAC1_MOUSE | O88942 | |
| NFAC2_MOUSE.H11DI.0.C |  |
MOUSE:Nfatc2 | 16 | C | 0 | nvvWGGAAAvdnhddn | HOCOMOCOv11 | ChIP-Seq | 0.954 | 4 | 507 | NFAT-related factors{6.1.3} | NFATc2 (NFATp, NFAT1){6.1.3.0.2} | 102463 | 18019 | NFAC2_MOUSE | Q60591 | ||
| NFE2_MOUSE.H11DI.0.A |  |
MOUSE:Nfe2 | 17 | A | 0 | vRTGACTCAGCAndhhn | HOCOMOCOv11 | ChIP-Seq | 0.904 | 0.996 | 17 | 9 | 392 | Jun-related factors{1.1.1} | NF-E2-like factors{1.1.1.2} | 97308 | 18022 | NFE2_MOUSE | Q07279 |
| NFIB_MOUSE.H11DI.0.C |  |
MOUSE:Nfib | 13 | C | 0 | nSYTGGCWbhndn | HOCOMOCOv11 | ChIP-Seq | 0.624 | 0.845 | 2 | 2 | 501 | Nuclear factor 1{7.1.2} | NF-1B{7.1.2.0.2} | 103188 | 18028 | NFIB_MOUSE | P97863 |
| NFIC_MOUSE.H11DI.0.A |  |
MOUSE:Nfic | 17 | A | 0 | nbYTGGCWbnndbYYhd | HOCOMOCOv11 | ChIP-Seq | 0.976 | 0.879 | 18 | 1 | 295 | Nuclear factor 1{7.1.2} | NF-1C{7.1.2.0.3} | 109591 | 18029 | NFIC_MOUSE | P70255 |
| NFIL3_MOUSE.H11DI.0.C |  |
MOUSE:Nfil3 | 12 | C | 0 | dvTTATGYAAbn | HOCOMOCOv11 | ChIP-Seq | 0.576 | 0.878 | 2 | 2 | 504 | C/EBP-related{1.1.8} | PAR factors{1.1.8.2} | 109495 | 18030 | NFIL3_MOUSE | O08750 |
| NFKB1_MOUSE.H11DI.0.A |  |
MOUSE:Nfkb1 | 14 | A | 0 | ndGGGRAAbhCCCh | HOCOMOCOv11 | ChIP-Seq | 0.787 | 0.965 | 7 | 16 | 528 | NF-kappaB-related factors{6.1.1} | NF-kappaB p50 subunit-like factors{6.1.1.1} | 97312 | 18033 | NFKB1_MOUSE | P25799 |
| NFYA_MOUSE.H11DI.0.A |  |
MOUSE:Nfya | 16 | A | 0 | nnbRdCCAATvRvMRn | HOCOMOCOv11 | ChIP-Seq | 0.983 | 0.957 | 19 | 14 | 507 | Heteromeric CCAAT-binding factors{4.2.1} | NF-YA (CP1A, CBF-B){4.2.1.0.1} | 97316 | 18044 | NFYA_MOUSE | P23708 |
| NFYB_MOUSE.H11DI.0.A | 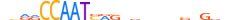 |
MOUSE:Nfyb | 23 | A | 0 | bbRRCCAATSRSvdbbvnvnSbv | HOCOMOCOv10 | ChIP-Seq | 0.987 | 0.998 | 4 | 4 | 490 | Heteromeric CCAAT-binding factors{4.2.1} | NF-YB (CP1B, CBF-A){4.2.1.0.2} | 97317 | 18045 | NFYB_MOUSE | P63139 |
| NFYC_MOUSE.H11DI.0.A |  |
MOUSE:Nfyc | 17 | A | 0 | nnbbRRCCAATSASvRn | HOCOMOCOv11 | ChIP-Seq | 0.844 | 0.989 | 4 | 4 | 507 | Heteromeric CCAAT-binding factors{4.2.1} | NF-YC{4.2.1.0.3} | 107901 | 18046 | NFYC_MOUSE | P70353 |
| NGN2_MOUSE.H11DI.0.C |  |
MOUSE:Neurog2 | 13 | C | 0 | vvnRRCAGATGGb | HOCOMOCOv11 | ChIP-Seq | 0.984 | 4 | 513 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 109619 | 11924 | NGN2_MOUSE | P70447 | ||
| NKX21_MOUSE.H11DI.0.A | 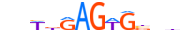 |
MOUSE:Nkx2-1 | 13 | A | 0 | nbWbRAGWGbbhn | HOCOMOCOv11 | ChIP-Seq | 0.905 | 0.937 | 14 | 9 | 502 | NK-related factors{3.1.2} | NK-2.1{3.1.2.14} | 108067 | 21869 | NKX21_MOUSE | P50220 |
| NKX22_MOUSE.H11DI.0.A |  |
MOUSE:Nkx2-2 | 10 | A | 0 | nYbRAGTGbn | HOCOMOCOv11 | ChIP-Seq | 0.389 | 0.818 | 2 | 5 | 510 | NK-related factors{3.1.2} | NK-2.2{3.1.2.15} | 97347 | 18088 | NKX22_MOUSE | P42586 |
| NKX25_MOUSE.H11DI.0.B |  |
MOUSE:Nkx2-5 | 11 | B | 0 | nbhKGAGTGbn | HOCOMOCOv11 | ChIP-Seq | 0.662 | 0.882 | 2 | 7 | 525 | NK-related factors{3.1.2} | NK-4{3.1.2.17} | 97350 | 18091 | NKX25_MOUSE | P42582 |
| NKX31_MOUSE.H11DI.0.B |  |
MOUSE:Nkx3-1 | 11 | B | 0 | nbWRWGTRYYh | HOCOMOCOv11 | ChIP-Seq | 0.844 | 0.798 | 11 | 10 | 427 | NK-related factors{3.1.2} | NK-3{3.1.2.16} | 97352 | 18095 | NKX31_MOUSE | P97436 |
| NKX32_MOUSE.H11DI.0.B |  |
MOUSE:Nkx3-2 | 12 | B | 0 | nWTAAGTGbdYn | HOCOMOCOv11 | ChIP-Seq | 0.979 | 6 | 585 | NK-related factors{3.1.2} | NK-3{3.1.2.16} | 108015 | 12020 | NKX32_MOUSE | P97503 | ||
| NKX61_MOUSE.H11DI.0.A |  |
MOUSE:Nkx6-1 | 22 | A | 0 | ddRhdRAKKdbhdWWWRAWddn | HOCOMOCOv11 | ChIP-Seq | 0.435 | 0.975 | 3 | 8 | 510 | NK-related factors{3.1.2} | NK-6{3.1.2.19} | 1206039 | 18096 | NKX61_MOUSE | Q99MA9 |
| NR1D1_MOUSE.H11DI.0.A |  |
MOUSE:Nr1d1 | 19 | A | 0 | nvnvWbKRGGbCAvnndvn | HOCOMOCOv11 | ChIP-Seq | 0.59 | 0.907 | 1 | 53 | 508 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Rev-ErbA (NR1D){2.1.2.3} | 2444210 | 217166 | NR1D1_MOUSE | Q3UV55 |
| NR1D2_MOUSE.H11DI.0.A |  |
MOUSE:Nr1d2 | 18 | A | 0 | nvvdvWvTRRGTCAbnvn | HOCOMOCOv11 | ChIP-Seq | 0.898 | 12 | 511 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Rev-ErbA (NR1D){2.1.2.3} | 2449205 | 353187 | NR1D2_MOUSE | Q60674 | ||
| NR1H3_MOUSE.H11DI.0.A |  |
MOUSE:Nr1h3 | 20 | A | 0 | nvdKbnRhhhvAGGdCAnnb | HOCOMOCOv11 | ChIP-Seq | 0.84 | 0.94 | 13 | 10 | 501 | Thyroid hormone receptor-related factors (NR1){2.1.2} | LXR (NR1H){2.1.2.7} | 1352462 | 22259 | NR1H3_MOUSE | Q9Z0Y9 |
| NR1H4_MOUSE.H11DI.0.A |  |
MOUSE:Nr1h4 | 22 | A | 0 | vRGKKSRvYGdCCbvbvKKbYv | HOCOMOCOv11 | ChIP-Seq | 0.942 | 15 | 472 | Thyroid hormone receptor-related factors (NR1){2.1.2} | LXR (NR1H){2.1.2.7} | 1352464 | 20186 | NR1H4_MOUSE | Q60641 | ||
| NR4A1_MOUSE.H11DI.0.B |  |
MOUSE:Nr4a1 | 10 | B | 0 | nAAAKGYCAn | HOCOMOCOv11 | ChIP-Seq | 0.929 | 0.586 | 12 | 2 | 511 | NGFI-B-related receptors (NR4){2.1.4} | NGFI-B (Nur77, NR4A1){2.1.4.0.1} | 1352454 | 15370 | NR4A1_MOUSE | P12813 |
| NR5A2_MOUSE.H11DI.0.A |  |
MOUSE:Nr5a2 | 16 | A | 0 | ndbYCAAGGYCRnnnn | HOCOMOCOv10 | ChIP-Seq | 0.526 | 0.957 | 4 | 8 | 501 | FTZ-F1-related receptors (NR5){2.1.5} | LRH-1 (NR5A2){2.1.5.0.2} | 1346834 | 26424 | NR5A2_MOUSE | P45448 |
| NR6A1_MOUSE.H11DI.0.C |  |
MOUSE:Nr6a1 | 16 | C | 0 | nvdddbYCAWRKbYRn | HOCOMOCOv11 | ChIP-Seq | 0.802 | 5 | 505 | GCNF-related receptors (NR6){2.1.6} | GCNF (NR6A1){2.1.6.0.1} | 1352459 | 14536 | NR6A1_MOUSE | Q64249 | ||
| NRF1_MOUSE.H11DI.0.A |  |
MOUSE:Nrf1 | 20 | A | 0 | nnnnbvnbGCGCAKGCGCRv | HOCOMOCOv11 | ChIP-Seq | 0.999 | 0.999 | 31 | 23 | 407 | NRF{0.0.6} | NRF-1 (alpha-pal){0.0.6.0.1} | 1332235 | 18181 | NRF1_MOUSE | Q9WU00 |
| OLIG2_MOUSE.H11DI.0.A | 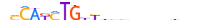 |
MOUSE:Olig2 | 22 | A | 0 | nMCAKMTGYYbhbndbhhbnhn | HOCOMOCOv11 | ChIP-Seq | 0.941 | 19 | 500 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 1355331 | 50913 | OLIG2_MOUSE | Q9EQW6 | ||
| OTX2_MOUSE.H11DI.0.A |  |
MOUSE:Otx2 | 14 | A | 0 | ndddRGGATTAdvn | HOCOMOCOv11 | ChIP-Seq | 0.879 | 0.92 | 5 | 18 | 500 | Paired-related HD factors{3.1.3} | OTX{3.1.3.17} | 97451 | OTX2_MOUSE | P80206 | |
| OVOL2_MOUSE.H11DI.0.C |  |
MOUSE:Ovol2 | 11 | C | 0 | ndndTAACGGd | HOCOMOCOv11 | ChIP-Seq | 0.454 | 0.963 | 2 | 2 | 185 | More than 3 adjacent zinc finger factors{2.3.3} | OVOL-factors{2.3.3.17} | 1338039 | 107586 | OVOL2_MOUSE | Q8CIV7 |
| P53_MOUSE.H11DI.0.A |  |
MOUSE:Tp53 | 23 | A | 0 | vnRRACAWGYMYdGRCWdGhYhn | HOCOMOCOv11 | ChIP-Seq | 0.999 | 0.994 | 141 | 39 | 514 | p53-related factors{6.3.1} | p53{6.3.1.0.1} | 98834 | 22059 | P53_MOUSE | P02340 |
| PAX3_MOUSE.H11DI.0.C |  |
MOUSE:Pax3 | 16 | C | 0 | nnbvdnWAATbdvddd | HOCOMOCOv11 | ChIP-Seq | 0.817 | 3 | 447 | Paired plus homeo domain{3.2.1} | PAX-3/7{3.2.1.1} | 97487 | 18505 | PAX3_MOUSE | P24610 | ||
| PAX5_MOUSE.H11DI.0.A |  |
MOUSE:Pax5 | 20 | A | 0 | nddbYvvvvRRSCRKGRMbn | HOCOMOCOv11 | ChIP-Seq | 0.947 | 0.927 | 32 | 8 | 401 | Paired domain only{3.2.2} | PAX-2-like factors (partial homeobox){3.2.2.2} | 97489 | 18507 | PAX5_MOUSE | Q02650 |
| PAX6_MOUSE.H11DI.0.B |  |
MOUSE:Pax6 | 15 | B | 0 | hKYvYRChTSRnYRv | HOCOMOCOv11 | ChIP-Seq | 0.606 | 0.8 | 6 | 8 | 500 | Paired plus homeo domain{3.2.1} | PAX-4/6{3.2.1.2} | 97490 | 18508 | PAX6_MOUSE | P63015 |
| PBX1_MOUSE.H11DI.0.A |  |
MOUSE:Pbx1 | 12 | A | 0 | nWGAYKGRYRRv | HOCOMOCOv11 | ChIP-Seq | 0.851 | 0.915 | 2 | 36 | 508 | TALE-type homeo domain factors{3.1.4} | PBX{3.1.4.4} | 97495 | 18514 | PBX1_MOUSE | P41778 |
| PDX1_MOUSE.H11DI.0.A |  |
MOUSE:Pdx1 | 12 | A | 0 | dTGAYWRAKKdn | HOCOMOCOv11 | ChIP-Seq | 0.949 | 0.905 | 10 | 4 | 503 | HOX-related factors{3.1.1} | PDX{3.1.1.15} | 102851 | 18609 | PDX1_MOUSE | P52946 |
| PITX1_MOUSE.H11DI.0.C |  |
MOUSE:Pitx1 | 11 | C | 0 | nvdGGMTTAdn | HOCOMOCOv11 | ChIP-Seq | 0.523 | 0.887 | 2 | 4 | 390 | Paired-related HD factors{3.1.3} | PITX{3.1.3.19} | 107374 | 18740 | PITX1_MOUSE | P70314 |
| PKNX1_MOUSE.H11DI.0.A |  |
MOUSE:Pknox1 | 12 | A | 0 | bWGAbKGRCRKv | HOCOMOCOv11 | ChIP-Seq | 0.973 | 19 | 479 | TALE-type homeo domain factors{3.1.4} | PKNOX{3.1.4.5} | 1201409 | 18771 | PKNX1_MOUSE | O70477 | ||
| PO2F1_MOUSE.H11DI.0.B |  |
MOUSE:Pou2f1 | 19 | B | 0 | nbnYATGYAdATdhvnndn | HOCOMOCOv11 | ChIP-Seq | 0.822 | 0.687 | 15 | 14 | 103 | POU domain factors{3.1.10} | POU2 (Oct-1/2-like factors){3.1.10.2} | 101898 | 18986 | PO2F1_MOUSE | P25425 |
| PO3F1_MOUSE.H11DI.0.B |  |
MOUSE:Pou3f1 | 19 | B | 0 | nbdbWTGMATWhnnMWdvn | HOCOMOCOv11 | ChIP-Seq | 0.849 | 10 | 504 | POU domain factors{3.1.10} | POU3 (Oct-6-like factors){3.1.10.3} | 101896 | 18991 | PO3F1_MOUSE | P21952 | ||
| PO3F2_MOUSE.H11DI.0.A |  |
MOUSE:Pou3f2 | 19 | A | 0 | bdnnbdYATGMATAWKYMW | HOCOMOCOv11 | ChIP-Seq | 0.787 | 0.956 | 12 | 22 | 385 | POU domain factors{3.1.10} | POU3 (Oct-6-like factors){3.1.10.3} | 101895 | 18992 | PO3F2_MOUSE | P31360 |
| PO5F1_MOUSE.H11DI.0.A |  |
MOUSE:Pou5f1 | 19 | A | 0 | bbhWTTSThATGYWRAWdn | HOCOMOCOv11 | ChIP-Seq | 0.984 | 0.988 | 44 | 119 | 495 | POU domain factors{3.1.10} | POU5 (Oct-3/4-like factors){3.1.10.5} | 101893 | 18999 | PO5F1_MOUSE | P20263 |
| PPARA_MOUSE.H11DI.0.A |  |
MOUSE:Ppara | 19 | A | 0 | nhWbYRKGbbARAGGKbMn | HOCOMOCOv11 | ChIP-Seq | 0.59 | 0.965 | 5 | 20 | 500 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 104740 | 19013 | PPARA_MOUSE | P23204 |
| PPARG_MOUSE.H11DI.0.A |  |
MOUSE:Pparg | 20 | A | 0 | vnhdbbRKGbSARAGGKbRn | HOCOMOCOv11 | ChIP-Seq | 0.969 | 0.975 | 53 | 114 | 498 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 97747 | 19016 | PPARG_MOUSE | P37238 |
| PRD14_MOUSE.H11DI.0.A |  |
MOUSE:Prdm14 | 18 | A | 0 | ndddSKTAGRGMMSbddn | HOCOMOCOv11 | ChIP-Seq | 0.881 | 0.947 | 7 | 11 | 535 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 3588194 | 383491 | PRD14_MOUSE | E9Q3T6 |
| PRD16_MOUSE.H11DI.0.B |  |
MOUSE:Prdm16 | 13 | B | 0 | nKCCCWRRRGRvn | HOCOMOCOv11 | ChIP-Seq | 0.876 | 3 | 501 | Factors with multiple dispersed zinc fingers{2.3.4} | Evi-1-like factors{2.3.4.14} | 1917923 | 70673 | PRD16_MOUSE | A2A935 | ||
| PRDM1_MOUSE.H11DI.0.A |  |
MOUSE:Prdm1 | 17 | A | 0 | ddRMAGKGAAAGTdhnn | HOCOMOCOv10 | ChIP-Seq | 0.982 | 0.959 | 12 | 9 | 501 | More than 3 adjacent zinc finger factors{2.3.3} | PRDM1-like factors{2.3.3.12} | 99655 | 12142 | PRDM1_MOUSE | Q60636 |
| PRDM5_MOUSE.H11DI.0.A |  |
MOUSE:Prdm5 | 19 | A | 0 | nhnvvnGGWGvdSvRGKhn | HOCOMOCOv11 | ChIP-Seq | 0.956 | 12 | 507 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 1918029 | 70779 | PRDM5_MOUSE | Q9CXE0 | ||
| PRDM9_MOUSE.H11DI.0.C |  |
MOUSE:Prdm9 | 21 | C | 0 | nhdnddRRYAGYAvCWbChbv | HOCOMOCOv11 | ChIP-Seq | 0.91 | 4 | 245 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 2384854 | PRDM9_MOUSE | Q96EQ9 | |||
| PRGR_MOUSE.H11DI.0.A |  |
MOUSE:Pgr | 16 | A | 0 | nRGnACRnRvWGTnCh | HOCOMOCOv11 | ChIP-Seq | 0.964 | 0.985 | 51 | 15 | 405 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 97567 | PRGR_MOUSE | Q00175 | |
| PROP1_MOUSE.H11DI.0.C |  |
MOUSE:Prop1 | 13 | C | 0 | nWAATKAvRTTWn | HOCOMOCOv11 | ChIP-Seq | 0.928 | 3 | 501 | Paired-related HD factors{3.1.3} | PROP{3.1.3.20} | 109330 | 19127 | PROP1_MOUSE | P97458 | ||
| PTF1A_MOUSE.H11DI.0.A | 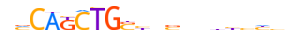 |
MOUSE:Ptf1a | 20 | A | 0 | nMCAKCTGSbnbhbhbbhhn | HOCOMOCOv11 | ChIP-Seq | 0.969 | 20 | 500 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 1328312 | 19213 | PTF1A_MOUSE | Q9QX98 | ||
| RARA_MOUSE.H11DI.0.A |  |
MOUSE:Rara | 23 | A | 0 | vddvdbbvdRAGKTCARRKbYRb | HOCOMOCOv11 | ChIP-Seq | 0.863 | 0.945 | 29 | 32 | 498 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 97856 | 19401 | RARA_MOUSE | P11416 |
| REST_MOUSE.H11DI.0.A |  |
MOUSE:Rest | 24 | A | 0 | nYCAGSACCdhGGACAGMdSYhnv | HOCOMOCOv11 | ChIP-Seq | 0.999 | 0.998 | 147 | 33 | 332 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 104897 | 19712 | REST_MOUSE | Q8VIG1 |
| RFX1_MOUSE.H11DI.0.A |  |
MOUSE:Rfx1 | 23 | A | 0 | bbbbSbGTTGCCWdGGvvMSvvb | HOCOMOCOv10 | ChIP-Seq | 0.699 | 0.994 | 3 | 7 | 440 | RFX-related factors{3.3.3} | RFX1 (EF-C){3.3.3.0.1} | 105982 | 19724 | RFX1_MOUSE | P48377 |
| RFX2_MOUSE.H11DI.0.A |  |
MOUSE:Rfx2 | 20 | A | 0 | bbbGTTGCCWKGGhRACvvv | HOCOMOCOv11 | ChIP-Seq | 0.978 | 0.998 | 7 | 7 | 360 | RFX-related factors{3.3.3} | RFX2{3.3.3.0.2} | 106583 | 19725 | RFX2_MOUSE | P48379 |
| RFX3_MOUSE.H11DI.0.C |  |
MOUSE:Rfx3 | 18 | C | 0 | bSbGTTGCCWWGGnRACv | HOCOMOCOv11 | ChIP-Seq | 0.994 | 2 | 354 | RFX-related factors{3.3.3} | RFX3{3.3.3.0.3} | 106582 | 19726 | RFX3_MOUSE | P48381 | ||
| RFX6_MOUSE.H11DI.0.C |  |
MOUSE:Rfx6 | 16 | C | 0 | bbGYWKCYvdGnnvvn | HOCOMOCOv11 | ChIP-Seq | 0.954 | 3 | 522 | RFX-related factors{3.3.3} | RFX6 (RFXDC-1){3.3.3.0.6} | 2445208 | 320995 | RFX6_MOUSE | Q8C7R7 | ||
| RORA_MOUSE.H11DI.0.B |  |
MOUSE:Rora | 16 | B | 0 | ndvhndRGGYCAvnvn | HOCOMOCOv11 | ChIP-Seq | 0.58 | 0.809 | 2 | 5 | 501 | Thyroid hormone receptor-related factors (NR1){2.1.2} | ROR (NR1F){2.1.2.6} | 104661 | 19883 | RORA_MOUSE | P51448 |
| RORG_MOUSE.H11DI.0.B |  |
MOUSE:Rorc | 13 | B | 0 | ndWWvTRRGTCAn | HOCOMOCOv11 | ChIP-Seq | 0.928 | 19 | 504 | Thyroid hormone receptor-related factors (NR1){2.1.2} | ROR (NR1F){2.1.2.6} | 104856 | 19885 | RORG_MOUSE | P51450 | ||
| RUNX1_MOUSE.H11DI.0.A | 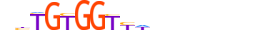 |
MOUSE:Runx1 | 17 | A | 0 | nbTGTGGTYWbnbbnbn | HOCOMOCOv11 | ChIP-Seq | 0.966 | 0.985 | 115 | 63 | 514 | Runt-related factors{6.4.1} | Runx1 (PEBP2alphaB, CBF-alpha2, AML-1){6.4.1.0.2} | 99852 | RUNX1_MOUSE | Q03347 | |
| RUNX2_MOUSE.H11DI.0.A |  |
MOUSE:Runx2 | 13 | A | 0 | nbYTGTGGTYdbn | HOCOMOCOv11 | ChIP-Seq | 0.856 | 0.937 | 3 | 25 | 547 | Runt-related factors{6.4.1} | Runx2 (PEBP2alphaA, CBF-alpha1){6.4.1.0.1} | 99829 | 12393 | RUNX2_MOUSE | Q08775 |
| RUNX3_MOUSE.H11DI.0.A |  |
MOUSE:Runx3 | 15 | A | 0 | bnhTGTGGTbdnbvn | HOCOMOCOv11 | ChIP-Seq | 0.994 | 0.979 | 2 | 27 | 198 | Runt-related factors{6.4.1} | Runx3 (PEBP2alphaC, CBF-alpha3, AML-2){6.4.1.0.3} | 102672 | RUNX3_MOUSE | Q64131 | |
| RXRA_MOUSE.H11DI.0.A |  |
MOUSE:Rxra | 25 | A | 0 | dRRddvddbvdvAGKTCAvRRhvdn | HOCOMOCOv11 | ChIP-Seq | 0.772 | 0.988 | 52 | 158 | 503 | RXR-related receptors (NR2){2.1.3} | Retinoid X receptors (NR2B){2.1.3.1} | 98214 | 20181 | RXRA_MOUSE | P28700 |
| RXRG_MOUSE.H11DI.0.A |  |
MOUSE:Rxrg | 25 | A | 0 | dvddvdbnnnRRGKKSARGGYYRnv | HOCOMOCOv11 | ChIP-Seq | 0.986 | 7 | 395 | RXR-related receptors (NR2){2.1.3} | Retinoid X receptors (NR2B){2.1.3.1} | 98216 | 20183 | RXRG_MOUSE | P28705 | ||
| SALL1_MOUSE.H11DI.0.C |  |
MOUSE:Sall1 | 19 | C | 0 | nhhTYWRCAWhWvWRhdvn | HOCOMOCOv11 | ChIP-Seq | 0.801 | 3 | 499 | Factors with multiple dispersed zinc fingers{2.3.4} | Sal-like factors{2.3.4.3} | 1889585 | SALL1_MOUSE | Q9ER74 | |||
| SALL4_MOUSE.H11DI.0.A | 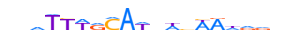 |
MOUSE:Sall4 | 19 | A | 0 | nnhTYWRMAWdWvWRdnvn | HOCOMOCOv11 | ChIP-Seq | 0.427 | 0.937 | 1 | 21 | 502 | Factors with multiple dispersed zinc fingers{2.3.4} | Sal-like factors{2.3.4.3} | 2139360 | 99377 | SALL4_MOUSE | Q8BX22 |
| SIX2_MOUSE.H11DI.0.A |  |
MOUSE:Six2 | 16 | A | 0 | ndGdAAbhhRAbMYbn | HOCOMOCOv11 | ChIP-Seq | 0.975 | 0.96 | 18 | 9 | 489 | HD-SINE factors{3.1.6} | SIX1-like factors{3.1.6.1} | 102778 | 20472 | SIX2_MOUSE | Q62232 |
| SIX4_MOUSE.H11DI.0.C |  |
MOUSE:Six4 | 15 | C | 0 | ndGdMAbhbRAbhbn | HOCOMOCOv11 | ChIP-Seq | 0.916 | 4 | 428 | HD-SINE factors{3.1.6} | SIX4-like factors{3.1.6.3} | 106034 | 20474 | SIX4_MOUSE | Q61321 | ||
| SMAD2_MOUSE.H11DI.0.A |  |
MOUSE:Smad2 | 14 | A | 0 | nbTGTYTGbbhhbn | HOCOMOCOv11 | ChIP-Seq | 0.849 | 0.901 | 17 | 26 | 501 | SMAD factors{7.1.1} | Regulatory Smads (R-Smad){7.1.1.1} | 108051 | 17126 | SMAD2_MOUSE | Q62432 |
| SMAD3_MOUSE.H11DI.0.C | 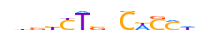 |
MOUSE:Smad3 | 14 | C | 0 | nhvbYTvbCWSMhn | HOCOMOCOv11 | ChIP-Seq | 0.799 | 0.682 | 34 | 30 | 506 | SMAD factors{7.1.1} | Regulatory Smads (R-Smad){7.1.1.1} | 1201674 | 17127 | SMAD3_MOUSE | Q8BUN5 |
| SMAD4_MOUSE.H11DI.0.A |  |
MOUSE:Smad4 | 21 | A | 0 | nnbWGKCTvdCWSYYnnnnnn | HOCOMOCOv10 | ChIP-Seq | 0.882 | 0.926 | 24 | 3 | 500 | SMAD factors{7.1.1} | Co-activating Smads (Co-Smad){7.1.1.2} | 894293 | 17128 | SMAD4_MOUSE | P97471 |
| SMCA5_MOUSE.H11DI.0.C | 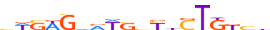 |
MOUSE:Smarca5 | 18 | C | 0 | hWSWGvMWSvYdSTKKYh | HOCOMOCOv11 | ChIP-Seq | 0.668 | 0.729 | 3 | 2 | 274 | Myb/SANT domain factors{3.5.1} | SMARCA-like factors{3.5.1.5} | 1935129 | 93762 | SMCA5_MOUSE | Q91ZW3 |
| SNAI2_MOUSE.H11DI.0.A |  |
MOUSE:Snai2 | 13 | A | 0 | ndRCAGGTGYnbb | HOCOMOCOv11 | ChIP-Seq | 0.976 | 0.94 | 11 | 3 | 417 | More than 3 adjacent zinc finger factors{2.3.3} | Snail-like factors{2.3.3.2} | 1096393 | 20583 | SNAI2_MOUSE | P97469 |
| SOX10_MOUSE.H11DI.0.B |  |
MOUSE:Sox10 | 19 | B | 0 | hvARdvvnnbCWTTGTbbn | HOCOMOCOv11 | ChIP-Seq | 0.936 | 0.781 | 3 | 3 | 502 | 98358 | 20665 | SOX10_MOUSE | Q04888 | ||
| SOX2_MOUSE.H11DI.0.A |  |
MOUSE:Sox2 | 19 | A | 0 | nbSYWTTGTbhWKbhnMhn | HOCOMOCOv11 | ChIP-Seq | 0.971 | 0.976 | 67 | 109 | 512 | SOX-related factors{4.1.1} | Group B{4.1.1.2} | 98364 | 20674 | SOX2_MOUSE | P48432 |
| SOX3_MOUSE.H11DI.0.C |  |
MOUSE:Sox3 | 13 | C | 0 | nbYCWTTGTbbbb | HOCOMOCOv11 | ChIP-Seq | 0.595 | 0.911 | 2 | 4 | 348 | SOX-related factors{4.1.1} | Group B{4.1.1.2} | 98365 | SOX3_MOUSE | P53784 | |
| SOX4_MOUSE.H11DI.0.A |  |
MOUSE:Sox4 | 13 | A | 0 | nbYCTTTGTYYbb | HOCOMOCOv11 | ChIP-Seq | 0.637 | 0.981 | 2 | 7 | 500 | SOX-related factors{4.1.1} | Group C{4.1.1.3} | 98366 | 20677 | SOX4_MOUSE | Q06831 |
| SOX9_MOUSE.H11DI.0.A |  |
MOUSE:Sox9 | 19 | A | 0 | vMMAAdRvvnbCWTTGTKb | HOCOMOCOv11 | ChIP-Seq | 0.655 | 0.989 | 6 | 25 | 500 | SOX-related factors{4.1.1} | Group E{4.1.1.5} | 98371 | 20682 | SOX9_MOUSE | Q04887 |
| SP1_MOUSE.H11DI.0.A |  |
MOUSE:Sp1 | 27 | A | 0 | vSSvRRGGGMGGGGvvdRRRSvSRvRv | HOCOMOCOv11 | ChIP-Seq | 0.999 | 0.991 | 53 | 11 | 494 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 98372 | 20683 | SP1_MOUSE | O89090 |
| SP5_MOUSE.H11DI.0.C | 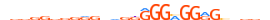 |
MOUSE:Sp5 | 27 | C | 0 | dddRRvvRRRvdRKGGGMGGRGhdvvd | HOCOMOCOv11 | ChIP-Seq | 0.966 | 3 | 511 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 1927715 | 64406 | SP5_MOUSE | Q9JHX2 | ||
| SP7_MOUSE.H11DI.0.A |  |
MOUSE:Sp7 | 16 | A | 0 | vhRvSMWvYMATTWnn | HOCOMOCOv11 | ChIP-Seq | 0.979 | 19 | 410 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 2153568 | 170574 | SP7_MOUSE | Q8VI67 | ||
| SPI1_MOUSE.H11DI.0.A |  |
MOUSE:Spi1 | 18 | A | 0 | nvvRRWGRGGAAGTGvnv | HOCOMOCOv11 | ChIP-Seq | 0.999 | 0.999 | 80 | 204 | 505 | Ets-related factors{3.5.2} | Spi-like factors{3.5.2.5} | 98282 | 20375 | SPI1_MOUSE | P17433 |
| SPIB_MOUSE.H11DI.0.A |  |
MOUSE:Spib | 19 | A | 0 | nvRRWRRGGAASTGRdRvn | HOCOMOCOv11 | ChIP-Seq | 0.999 | 0.983 | 12 | 3 | 500 | Ets-related factors{3.5.2} | Spi-like factors{3.5.2.5} | 892986 | 272382 | SPIB_MOUSE | O35906 |
| SRBP1_MOUSE.H11DI.0.B | 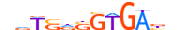 |
MOUSE:Srebf1 | 12 | B | 0 | nvTSRSGTGAbn | HOCOMOCOv11 | ChIP-Seq | 0.919 | 0.699 | 5 | 23 | 603 | bHLH-ZIP factors{1.2.6} | SREBP factors{1.2.6.3} | 107606 | 20787 | SRBP1_MOUSE | Q9WTN3 |
| SRF_MOUSE.H11DI.0.A |  |
MOUSE:Srf | 21 | A | 0 | hhhdYKbCCTTWTWTGGvhdb | HOCOMOCOv11 | ChIP-Seq | 0.951 | 0.988 | 46 | 37 | 322 | Responders to external signals (SRF/RLM1){5.1.2} | SRF{5.1.2.0.1} | 106658 | 20807 | SRF_MOUSE | Q9JM73 |
| STA5A_MOUSE.H11DI.0.A |  |
MOUSE:Stat5a | 13 | A | 0 | ndnTTCYMRGAAn | HOCOMOCOv11 | ChIP-Seq | 0.927 | 0.993 | 24 | 127 | 500 | STAT factors{6.2.1} | STAT5A{6.2.1.0.5} | 103036 | 20850 | STA5A_MOUSE | P42230 |
| STA5B_MOUSE.H11DI.0.A |  |
MOUSE:Stat5b | 13 | A | 0 | nnTTCYTRGAAvn | HOCOMOCOv11 | ChIP-Seq | 0.966 | 0.965 | 16 | 31 | 501 | STAT factors{6.2.1} | STAT5B{6.2.1.0.6} | 103035 | 20851 | STA5B_MOUSE | P42232 |
| STAT1_MOUSE.H11DI.0.A |  |
MOUSE:Stat1 | 24 | A | 0 | ndRRRAAhhGAAASWvRWdvnvdn | HOCOMOCOv11 | ChIP-Seq | 0.988 | 0.966 | 37 | 128 | 506 | STAT factors{6.2.1} | STAT1{6.2.1.0.1} | 103063 | STAT1_MOUSE | P42225 | |
| STAT2_MOUSE.H11DI.0.A |  |
MOUSE:Stat2 | 21 | A | 0 | nvvRRAAhnGAAASYRRRdnn | HOCOMOCOv11 | ChIP-Seq | 0.981 | 0.994 | 11 | 8 | 509 | STAT factors{6.2.1} | STAT2{6.2.1.0.2} | 103039 | STAT2_MOUSE | Q9WVL2 | |
| STAT3_MOUSE.H11DI.0.A |  |
MOUSE:Stat3 | 20 | A | 0 | nhbvbTTMCYRKvAnbnnbn | HOCOMOCOv11 | ChIP-Seq | 0.95 | 0.946 | 36 | 82 | 509 | STAT factors{6.2.1} | STAT3{6.2.1.0.3} | 103038 | 20848 | STAT3_MOUSE | P42227 |
| STAT4_MOUSE.H11DI.0.A |  |
MOUSE:Stat4 | 22 | A | 0 | nvnnnbbnbTTCCYdKMAdnnn | HOCOMOCOv10 | ChIP-Seq | 0.958 | 0.942 | 7 | 10 | 502 | STAT factors{6.2.1} | STAT4{6.2.1.0.4} | 103062 | 20849 | STAT4_MOUSE | P42228 |
| STAT6_MOUSE.H11DI.0.A |  |
MOUSE:Stat6 | 18 | A | 0 | nbdnTTCYWvRGAAnnnn | HOCOMOCOv11 | ChIP-Seq | 0.771 | 0.951 | 19 | 24 | 492 | STAT factors{6.2.1} | STAT6{6.2.1.0.7} | 103034 | 20852 | STAT6_MOUSE | P52633 |
| STF1_MOUSE.H11DI.0.B |  |
MOUSE:Nr5a1 | 13 | B | 0 | nbbYCAAGGYCRn | HOCOMOCOv11 | ChIP-Seq | 0.775 | 0.908 | 1 | 2 | 500 | FTZ-F1-related receptors (NR5){2.1.5} | FTZ-F1 (SF-1) (NR5A1){2.1.5.0.1} | 1346833 | 26423 | STF1_MOUSE | P33242 |
| SUH_MOUSE.H11DI.0.A |  |
MOUSE:Rbpj | 10 | A | 0 | bbSTGRGAAh | HOCOMOCOv11 | ChIP-Seq | 0.952 | 0.958 | 25 | 6 | 510 | CSL-related factors{6.1.4} | M{6.1.4.1} | 96522 | 19664 | SUH_MOUSE | P31266 |
| TAF1_MOUSE.H11DI.0.A |  |
MOUSE:Taf1 | 18 | A | 0 | vRvvRWGGMRGMvvvvvn | HOCOMOCOv11 | ChIP-Seq | 0.846 | 0.919 | 61 | 2 | 500 | TCF-7-related factors{4.1.3} | TAF-1 (TAF-2A, TAF(II)250) [1]{4.1.3.0.5} | 1336878 | 270627 | TAF1_MOUSE | Q80UV9 |
| TAL1_MOUSE.H11DI.0.A |  |
MOUSE:Tal1 | 23 | A | 0 | nnnbbKbnnnnndvWGATAARvn | HOCOMOCOv10 | ChIP-Seq | 0.984 | 0.987 | 68 | 76 | 502 | Tal-related factors{1.2.3} | Tal / HEN-like factors{1.2.3.1} | 98480 | 21349 | TAL1_MOUSE | P22091 |
| TBP_MOUSE.H11DI.0.A |  |
MOUSE:Tbp | 13 | A | 0 | vWWKAAAWRvvvn | HOCOMOCOv11 | ChIP-Seq | 0.938 | 0.882 | 21 | 45 | 502 | TBP-related factors{8.1.1} | TBP{8.1.1.0.1} | 101838 | 21374 | TBP_MOUSE | P29037 |
| TBX20_MOUSE.H11DI.0.C |  |
MOUSE:Tbx20 | 14 | C | 0 | nvSTGnTGACARvn | HOCOMOCOv11 | ChIP-Seq | 0.91 | 4 | 468 | TBX1-related factors{6.5.3} | TBX20{6.5.3.0.5} | 1888496 | 57246 | TBX20_MOUSE | Q9ES03 | ||
| TBX21_MOUSE.H11DI.0.A |  |
MOUSE:Tbx21 | 16 | A | 0 | nbnddddSTGWSRvdn | HOCOMOCOv11 | ChIP-Seq | 0.71 | 0.877 | 7 | 17 | 488 | TBrain-related factors{6.5.2} | TBX21 (T-bet){6.5.2.0.3} | 1888984 | 57765 | TBX21_MOUSE | Q9JKD8 |
| TCF7_MOUSE.H11DI.0.A |  |
MOUSE:Tcf7 | 15 | A | 0 | nvvvASWWSAAARvn | HOCOMOCOv11 | ChIP-Seq | 0.858 | 0.944 | 7 | 19 | 499 | TCF-7-related factors{4.1.3} | TCF-7 (TCF-1) [1]{4.1.3.0.1} | 98507 | 21414 | TCF7_MOUSE | Q00417 |
| TEAD1_MOUSE.H11DI.0.A |  |
MOUSE:Tead1 | 13 | A | 0 | nRMATTCCWRSvn | HOCOMOCOv11 | ChIP-Seq | 0.961 | 0.952 | 14 | 5 | 439 | TEF-1-related factors{3.6.1} | TEF-1 (TEAD-1, TCF-13){3.6.1.0.1} | 101876 | 21676 | TEAD1_MOUSE | P30051 |
| TEAD2_MOUSE.H11DI.0.C |  |
MOUSE:Tead2 | 15 | C | 0 | nbRMATTCCWRSnhb | HOCOMOCOv11 | ChIP-Seq | 0.551 | 0.946 | 2 | 2 | 309 | TEF-1-related factors{3.6.1} | TEF-4 (TEAD-2){3.6.1.0.3} | 104904 | 21677 | TEAD2_MOUSE | P48301 |
| TEAD4_MOUSE.H11DI.0.A | 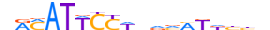 |
MOUSE:Tead4 | 18 | A | 0 | nRMATTCCWvdvMKdbbh | HOCOMOCOv11 | ChIP-Seq | 0.964 | 0.955 | 57 | 10 | 491 | TEF-1-related factors{3.6.1} | TEF-3 (TEAD-4, TCF-13L1, Tefr1){3.6.1.0.2} | 106907 | 21679 | TEAD4_MOUSE | Q62296 |
| TF2L1_MOUSE.H11DI.0.C | 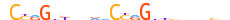 |
MOUSE:Tfcp2l1 | 23 | C | 0 | dCYRGhYbdddCYRGhYbnvnbh | HOCOMOCOv11 | ChIP-Seq | 0.995 | 3 | 454 | CP2-related factors{6.7.2} | CP2-L1 (LBP-9, CRTR-1){6.7.2.0.2} | 2444691 | 81879 | TF2L1_MOUSE | Q3UNW5 | ||
| TF65_MOUSE.H11DI.0.A | 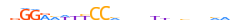 |
MOUSE:Rela | 24 | A | 0 | ndGGRMWKYCChvdvYbdbnhhMn | HOCOMOCOv11 | ChIP-Seq | 0.951 | 0.953 | 188 | 99 | 498 | NF-kappaB-related factors{6.1.1} | NF-kappaB p65 subunit-like factors{6.1.1.2} | 103290 | 19697 | TF65_MOUSE | Q04207 |
| TF7L1_MOUSE.H11DI.0.A |  |
MOUSE:Tcf7l1 | 13 | A | 0 | nbCTTTGAWvWbn | HOCOMOCOv11 | ChIP-Seq | 0.864 | 0.861 | 10 | 10 | 222 | TCF-7-related factors{4.1.3} | TCF-7L1 (TCF-3) [1]{4.1.3.0.2} | 1202876 | 21415 | TF7L1_MOUSE | Q9Z1J1 |
| TF7L2_MOUSE.H11DI.0.A |  |
MOUSE:Tcf7l2 | 16 | A | 0 | nvnnbbCTTTGAWSYb | HOCOMOCOv11 | ChIP-Seq | 0.958 | 0.911 | 46 | 10 | 500 | TCF-7-related factors{4.1.3} | TCF-7L2 (TCF-4) [1]{4.1.3.0.3} | 1202879 | 21416 | TF7L2_MOUSE | Q924A0 |
| TFE2_MOUSE.H11DI.0.A |  |
MOUSE:Tcf3 | 21 | A | 0 | nbnbbnbbhbnnvCASCTGYn | HOCOMOCOv11 | ChIP-Seq | 0.955 | 0.987 | 9 | 60 | 467 | E2A-related factors{1.2.1} | E2A (TCF-3, ITF-1){1.2.1.0.1} | 98510 | 21423 | TFE2_MOUSE | P15806 |
| TFE3_MOUSE.H11DI.0.A |  |
MOUSE:Tfe3 | 13 | A | 0 | ndGTCACGTGRbn | HOCOMOCOv11 | ChIP-Seq | 0.471 | 1.0 | 2 | 30 | 525 | bHLH-ZIP factors{1.2.6} | TFE3-like factors{1.2.6.1} | 98511 | 209446 | TFE3_MOUSE | Q64092 |
| TGIF1_MOUSE.H11DI.0.A |  |
MOUSE:Tgif1 | 11 | A | 0 | nTGACAGSnvn | HOCOMOCOv11 | ChIP-Seq | 0.812 | 0.981 | 2 | 5 | 142 | TALE-type homeo domain factors{3.1.4} | TGIF{3.1.4.6} | 1194497 | 21815 | TGIF1_MOUSE | P70284 |
| THA11_MOUSE.H11DI.0.B |  |
MOUSE:Thap11 | 25 | B | 0 | SCdnKGSMTbCTGGGAvdTGTAGTY | HOCOMOCOv11 | ChIP-Seq | 0.95 | 0.74 | 2 | 2 | 107 | THAP-related factors{2.9.1} | THAP11 (HRIHFB2206){2.9.1.0.11} | 1930964 | 59016 | THA11_MOUSE | Q9JJD0 |
| THA_MOUSE.H11DI.0.C |  |
MOUSE:Thra | 19 | C | 0 | nvRGKKSAnnYvAGGhCAn | HOCOMOCOv11 | ChIP-Seq | 0.946 | 4 | 505 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Thyroid hormone receptors (NR1A){2.1.2.2} | 98742 | 21833 | THA_MOUSE | P63058 | ||
| TWST1_MOUSE.H11DI.0.B |  |
MOUSE:Twist1 | 19 | B | 0 | nvCAKMTGKWWbbvATYWn | HOCOMOCOv11 | ChIP-Seq | 0.971 | 0.657 | 9 | 8 | 415 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 98872 | 22160 | TWST1_MOUSE | P26687 |
| TYY1_MOUSE.H11DI.0.A | 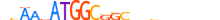 |
MOUSE:Yy1 | 22 | A | 0 | nvAAvATGGCSSCbvbvvbvbn | HOCOMOCOv10 | ChIP-Seq | 0.993 | 0.997 | 69 | 72 | 505 | More than 3 adjacent zinc finger factors{2.3.3} | YY1-like factors{2.3.3.9} | 99150 | 22632 | TYY1_MOUSE | Q00899 |
| USF1_MOUSE.H11DI.0.A |  |
MOUSE:Usf1 | 23 | A | 0 | vvbnvvnbbnvGTCACGTGvbbn | HOCOMOCOv10 | ChIP-Seq | 0.999 | 0.985 | 59 | 7 | 500 | bHLH-ZIP factors{1.2.6} | USF factors{1.2.6.2} | 99542 | 22278 | USF1_MOUSE | Q61069 |
| USF2_MOUSE.H11DI.0.A |  |
MOUSE:Usf2 | 23 | A | 0 | ndGTCACGTGvbbnvnnbnvnbb | HOCOMOCOv10 | ChIP-Seq | 0.998 | 0.999 | 23 | 10 | 517 | bHLH-ZIP factors{1.2.6} | USF factors{1.2.6.2} | 99961 | 22282 | USF2_MOUSE | Q64705 |
| VDR_MOUSE.H11DI.0.A |  |
MOUSE:Vdr | 18 | A | 0 | nvRGGKSAnnRRGKKCRn | HOCOMOCOv11 | ChIP-Seq | 0.953 | 0.939 | 48 | 30 | 499 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 103076 | 22337 | VDR_MOUSE | P48281 |
| VSX2_MOUSE.H11DI.0.C |  |
MOUSE:Vsx2 | 13 | C | 0 | nhTAATTAGCYdn | HOCOMOCOv11 | ChIP-Seq | 0.996 | 3 | 524 | Paired-related HD factors{3.1.3} | VSX{3.1.3.28} | 88401 | 12677 | VSX2_MOUSE | Q61412 | ||
| WT1_MOUSE.H11DI.0.A |  |
MOUSE:Wt1 | 23 | A | 0 | vvvGhGRRGGAGGRRvvRRvRRv | HOCOMOCOv11 | ChIP-Seq | 0.99 | 5 | 481 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 98968 | WT1_MOUSE | P22561 | |||
| ZBT17_MOUSE.H11DI.0.A |  |
MOUSE:Zbtb17 | 20 | A | 0 | vvvSSnGGRGGMGGGGvvvv | HOCOMOCOv11 | ChIP-Seq | 0.947 | 0.942 | 21 | 25 | 498 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 107410 | 22642 | ZBT17_MOUSE | Q60821 |
| ZFP42_MOUSE.H11DI.0.A |  |
MOUSE:Zfp42 | 15 | A | 0 | bRvhKCCATTYbdbd | HOCOMOCOv11 | ChIP-Seq | 0.938 | 0.84 | 3 | 2 | 343 | More than 3 adjacent zinc finger factors{2.3.3} | YY1-like factors{2.3.3.9} | 99187 | 22702 | ZFP42_MOUSE | P22227 |
| ZFX_MOUSE.H11DI.0.A |  |
MOUSE:Zfx | 12 | A | 0 | bYbvGGCCTdSn | HOCOMOCOv11 | ChIP-Seq | 0.84 | 0.993 | 10 | 4 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | ZFX/ZFY factors{2.3.3.65} | 99211 | 22764 | ZFX_MOUSE | P17012 |
| ZIC2_MOUSE.H11DI.0.C |  |
MOUSE:Zic2 | 17 | C | 0 | ndRhhhCCTGCTGdGhn | HOCOMOCOv11 | ChIP-Seq | 0.511 | 0.987 | 4 | 4 | 508 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 106679 | ZIC2_MOUSE | Q62520 | |
| ZIC3_MOUSE.H11DI.0.A |  |
MOUSE:Zic3 | 18 | A | 0 | nndShbhCCTGCTGdGhn | HOCOMOCOv11 | ChIP-Seq | 0.953 | 5 | 226 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 106676 | 22773 | ZIC3_MOUSE | Q62521 | ||
| ZKSC1_MOUSE.H11DI.0.A |  |
MOUSE:Zkscan1 | 21 | A | 0 | ndvhdbhCCTACTRWGTGYhn | HOCOMOCOv11 | ChIP-Seq | 0.969 | 6 | 476 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF24-like factors{2.3.3.10} | 1921820 | 74570 | ZKSC1_MOUSE | Q8BGS3 | ||
| ZN143_MOUSE.H11DI.0.A |  |
MOUSE:Znf143 | 25 | A | 0 | SCRvdGSMYbMTGGGAvdYGTAGTY | HOCOMOCOv11 | ChIP-Seq | 0.999 | 0.992 | 28 | 9 | 465 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF76-like factors{2.3.3.28} | 1277969 | 20841 | ZN143_MOUSE | O70230 |
| ZN281_MOUSE.H11DI.0.A |  |
MOUSE:Znf281 | 18 | A | 0 | vdbdGGGGGAGGGGvvvv | HOCOMOCOv11 | ChIP-Seq | 0.942 | 0.999 | 7 | 3 | 423 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF148-like factors{2.3.3.13} | 3029290 | 226442 | ZN281_MOUSE | Q99LI5 |
| ZN322_MOUSE.H11DI.0.B |  |
MOUSE:Znf322 | 22 | B | 0 | nSdKYCTRSYMCdSdGCMKSnv | HOCOMOCOv11 | ChIP-Seq | 0.905 | 0.699 | 4 | 2 | 426 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF322-like factors{2.3.3.52} | 2442566 | 218100 | ZN322_MOUSE | Q8BZ89 |
| ZN335_MOUSE.H11DI.0.A |  |
MOUSE:Znf335 | 25 | A | 0 | ndRGvYKbCYnvnRvdvYGCCTGWn | HOCOMOCOv11 | ChIP-Seq | 0.894 | 0.987 | 2 | 4 | 114 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 2682313 | 329559 | ZN335_MOUSE | A2A5K6 |
| ZN431_MOUSE.H11DI.0.C |  |
MOUSE:Znf431 | 27 | C | 0 | ndvWbRMYRACCTAAGACAGGvWbvvn | HOCOMOCOv11 | ChIP-Seq | 0.995 | 4 | 300 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF100-like factors{2.3.3.59} | 1916754 | 69504 | ZN431_MOUSE | E9QAG8 |