PWMs for HUMAN transcription factors (full)
| Model | LOGO | Transcription factor | Model length | Quality | Model rank | Consensus | Model release | Data source | Best auROC (human) | Best auROC (mouse) | Peak sets in benchmark (human) | Peak sets in benchmark (mouse) | Aligned words | TF family | TF subfamily | HGNC | EntrezGene | UniProt ID | UniProt AC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AHR_HUMAN.H11MO.0.B |  |
HUMAN:AHR | 9 | B | 0 | dKhGCGTGh | HOCOMOCOv9 | Integrative | 157 | PAS domain factors{1.2.5} | Ahr-like factors{1.2.5.1} | 348 | 196 | AHR_HUMAN | P35869 | ||||
| AIRE_HUMAN.H11MO.0.C |  |
HUMAN:AIRE | 18 | C | 0 | hnnGGWWnddWWGGdbWh | HOCOMOCOv9 | Integrative | 41 | AIRE{5.3.1} | AIRE{5.3.1.0.1} | 360 | 326 | AIRE_HUMAN | O43918 | ||||
| ALX1_HUMAN.H11MO.0.B |  |
HUMAN:ALX1 | 12 | B | 0 | vTRATYGnATTA | HOCOMOCOv9 | Integrative | 33 | Paired-related HD factors{3.1.3} | ALX{3.1.3.1} | 1494 | 8092 | ALX1_HUMAN | Q15699 | ||||
| ALX3_HUMAN.H11MO.0.D |  |
HUMAN:ALX3 | 11 | D | 0 | TAATTARMTWW | HOCOMOCOv10 | HT-SELEX | 8583 | Paired-related HD factors{3.1.3} | ALX{3.1.3.1} | 449 | 257 | ALX3_HUMAN | O95076 | ||||
| ALX4_HUMAN.H11MO.0.D |  |
HUMAN:ALX4 | 12 | D | 0 | hTAATTARATTA | HOCOMOCOv10 | HT-SELEX | 23663 | Paired-related HD factors{3.1.3} | ALX{3.1.3.1} | 450 | 60529 | ALX4_HUMAN | Q9H161 | ||||
| ANDR_HUMAN.H11MO.0.A |  |
HUMAN:AR | 18 | A | 0 | WKThYYddbhTRTTTRYh | HOCOMOCOv11 | ChIP-Seq | 0.96 | 0.811 | 461 | 57 | 499 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 644 | 367 | ANDR_HUMAN | P10275 |
| ANDR_HUMAN.H11MO.1.A |  |
HUMAN:AR | 16 | A | 1 | dGnACWbTbWGTdCYb | HOCOMOCOv11 | ChIP-Seq | 0.97 | 0.991 | 461 | 57 | 500 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 644 | 367 | ANDR_HUMAN | P10275 |
| ANDR_HUMAN.H11MO.2.A |  |
HUMAN:AR | 12 | A | 2 | nhbTGTKCYhdb | HOCOMOCOv11 | ChIP-Seq | 0.901 | 0.887 | 461 | 57 | 500 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 644 | 367 | ANDR_HUMAN | P10275 |
| AP2A_HUMAN.H11MO.0.A |  |
HUMAN:TFAP2A | 15 | A | 0 | dddSCCTGRGGShdd | HOCOMOCOv10 | ChIP-Seq | 0.956 | 0.937 | 13 | 13 | 500 | AP-2{1.3.1} | AP-2alpha{1.3.1.0.1} | 11742 | 7020 | AP2A_HUMAN | P05549 |
| AP2B_HUMAN.H11MO.0.B |  |
HUMAN:TFAP2B | 10 | B | 0 | GSCYGRGGRv | HOCOMOCOv9 | Integrative | 30 | AP-2{1.3.1} | AP-2beta{1.3.1.0.2} | 11743 | 7021 | AP2B_HUMAN | Q92481 | ||||
| AP2C_HUMAN.H11MO.0.A |  |
HUMAN:TFAP2C | 14 | A | 0 | vddSCCTGRGGShv | HOCOMOCOv10 | ChIP-Seq | 0.95 | 0.941 | 18 | 6 | 501 | AP-2{1.3.1} | AP-2gamma{1.3.1.0.3} | 11744 | 7022 | AP2C_HUMAN | Q92754 |
| AP2D_HUMAN.H11MO.0.D |  |
HUMAN:TFAP2D | 14 | D | 0 | nGbCCGRGGCnCGY | HOCOMOCOv9 | Integrative | 5 | AP-2{1.3.1} | AP-2delta{1.3.1.0.4} | 15581 | 83741 | AP2D_HUMAN | Q7Z6R9 | ||||
| ARI3A_HUMAN.H11MO.0.D |  |
HUMAN:ARID3A | 22 | D | 0 | dWTTAAWYvRdWWYAAAWYWWW | HOCOMOCOv9 | Integrative | 44 | ARID-related factors{3.7.1} | ARID3{3.7.1.3} | 3031 | 1820 | ARI3A_HUMAN | Q99856 | ||||
| ARI5B_HUMAN.H11MO.0.C |  |
HUMAN:ARID5B | 13 | C | 0 | nbYKnRTATTSKd | HOCOMOCOv9 | Integrative | 19 | ARID-related factors{3.7.1} | ARID5{3.7.1.5} | 17362 | 84159 | ARI5B_HUMAN | Q14865 | ||||
| ARNT2_HUMAN.H11MO.0.D |  |
HUMAN:ARNT2 | 12 | D | 0 | RnSGTGGvRSRS | HOCOMOCOv9 | Integrative | 18 | PAS domain factors{1.2.5} | Arnt-like factors{1.2.5.2} | 16876 | 9915 | ARNT2_HUMAN | Q9HBZ2 | ||||
| ARNT_HUMAN.H11MO.0.B |  |
HUMAN:ARNT | 9 | B | 0 | vdRCGTGMh | HOCOMOCOv11 | ChIP-Seq | 0.936 | 8 | 500 | PAS domain factors{1.2.5} | Arnt-like factors{1.2.5.2} | 700 | 405 | ARNT_HUMAN | P27540 | ||
| ARX_HUMAN.H11MO.0.D |  |
HUMAN:ARX | 13 | D | 0 | hTAATYTAATTWv | HOCOMOCOv10 | HT-SELEX | 623 | Paired-related HD factors{3.1.3} | ARX{3.1.3.3} | 18060 | 170302 | ARX_HUMAN | Q96QS3 | ||||
| ASCL1_HUMAN.H11MO.0.A |  |
HUMAN:ASCL1 | 14 | A | 0 | hnvCASCTGYYbhh | HOCOMOCOv11 | ChIP-Seq | 0.985 | 0.98 | 30 | 25 | 480 | MyoD / ASC-related factors{1.2.2} | Achaete-Scute-like factors{1.2.2.2} | 738 | 429 | ASCL1_HUMAN | P50553 |
| ASCL2_HUMAN.H11MO.0.D |  |
HUMAN:ASCL2 | 9 | D | 0 | vCASCTGCY | HOCOMOCOv11 | ChIP-Seq | 0.594 | 0.92 | 8 | 9 | 505 | MyoD / ASC-related factors{1.2.2} | Achaete-Scute-like factors{1.2.2.2} | 739 | 430 | ASCL2_HUMAN | Q99929 |
| ATF1_HUMAN.H11MO.0.B |  |
HUMAN:ATF1 | 11 | B | 0 | ndRTGACGYvd | HOCOMOCOv11 | ChIP-Seq | 0.892 | 6 | 500 | CREB-related factors{1.1.7} | CREB-like factors{1.1.7.1} | 783 | 466 | ATF1_HUMAN | P18846 | ||
| ATF2_HUMAN.H11MO.0.B |  |
HUMAN:ATF2 | 11 | B | 0 | dvTGAbGWMAb | HOCOMOCOv11 | ChIP-Seq | 0.751 | 0.849 | 8 | 4 | 502 | Jun-related factors{1.1.1} | ATF-2-like factors{1.1.1.3} | 784 | 1386 | ATF2_HUMAN | P15336 |
| ATF2_HUMAN.H11MO.1.B |  |
HUMAN:ATF2 | 20 | B | 1 | ddddRATGATGTCAThdhdd | HOCOMOCOv11 | ChIP-Seq | 0.705 | 0.806 | 8 | 4 | 1294 | Jun-related factors{1.1.1} | ATF-2-like factors{1.1.1.3} | 784 | 1386 | ATF2_HUMAN | P15336 |
| ATF2_HUMAN.H11MO.2.C |  |
HUMAN:ATF2 | 11 | C | 2 | RRTGACGTCAb | HOCOMOCOv11 | ChIP-Seq | 0.708 | 0.8 | 8 | 4 | 385 | Jun-related factors{1.1.1} | ATF-2-like factors{1.1.1.3} | 784 | 1386 | ATF2_HUMAN | P15336 |
| ATF3_HUMAN.H11MO.0.A |  |
HUMAN:ATF3 | 11 | A | 0 | dRTSACGTRRb | HOCOMOCOv11 | ChIP-Seq | 0.928 | 0.764 | 36 | 16 | 505 | Fos-related factors{1.1.2} | ATF-3-like factors{1.1.2.2} | 785 | 467 | ATF3_HUMAN | P18847 |
| ATF4_HUMAN.H11MO.0.A |  |
HUMAN:ATF4 | 12 | A | 0 | dnMTGATGCAAY | HOCOMOCOv11 | ChIP-Seq | 0.998 | 0.951 | 18 | 9 | 500 | ATF-4-related factors{1.1.6} | ATF-4{1.1.6.0.1} | 786 | 468 | ATF4_HUMAN | P18848 |
| ATF6A_HUMAN.H11MO.0.B |  |
HUMAN:ATF6 | 12 | B | 0 | vKSSTGACGTGG | HOCOMOCOv9 | Integrative | 20 | CREB-related factors{1.1.7} | ATF-6 factors{1.1.7.3} | 791 | 22926 | ATF6A_HUMAN | P18850 | ||||
| ATF7_HUMAN.H11MO.0.D |  |
HUMAN:ATF7 | 11 | D | 0 | dRTGACGTCAT | HOCOMOCOv10 | HT-SELEX | 13767 | Jun-related factors{1.1.1} | ATF-2-like factors{1.1.1.3} | 792 | 11016 | ATF7_HUMAN | P17544 | ||||
| ATOH1_HUMAN.H11MO.0.B |  |
HUMAN:ATOH1 | 9 | B | 0 | RRCAGMTGK | HOCOMOCOv11 | ChIP-Seq | 0.759 | 0.988 | 3 | 4 | 502 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 797 | 474 | ATOH1_HUMAN | Q92858 |
| BACH1_HUMAN.H11MO.0.A |  |
HUMAN:BACH1 | 13 | A | 0 | YGCTGAGTCMbbb | HOCOMOCOv10 | ChIP-Seq | 502 | Jun-related factors{1.1.1} | NF-E2-like factors{1.1.1.2} | 935 | 571 | BACH1_HUMAN | O14867 | ||||
| BACH2_HUMAN.H11MO.0.A |  |
HUMAN:BACH2 | 11 | A | 0 | WGCTKWSTCAb | HOCOMOCOv11 | ChIP-Seq | 0.926 | 0.98 | 10 | 3 | 500 | Jun-related factors{1.1.1} | NF-E2-like factors{1.1.1.2} | 14078 | 60468 | BACH2_HUMAN | Q9BYV9 |
| BARH1_HUMAN.H11MO.0.D |  |
HUMAN:BARHL1 | 13 | D | 0 | TAAWYGbYnhTWA | HOCOMOCOv10 | HT-SELEX | 5695 | NK-related factors{3.1.2} | BARHL{3.1.2.1} | 953 | 56751 | BARH1_HUMAN | Q9BZE3 | ||||
| BARH2_HUMAN.H11MO.0.D | 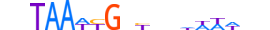 |
HUMAN:BARHL2 | 18 | D | 0 | vhTAAWYGnYbhhWWWhd | HOCOMOCOv10 | HT-SELEX | 41168 | NK-related factors{3.1.2} | BARHL{3.1.2.1} | 954 | 343472 | BARH2_HUMAN | Q9NY43 | ||||
| BARX1_HUMAN.H11MO.0.D |  |
HUMAN:BARX1 | 12 | D | 0 | nWMAKKSYTThh | HOCOMOCOv11 | ChIP-Seq | 0.608 | 0.831 | 2 | 2 | 461 | NK-related factors{3.1.2} | BARX{3.1.2.2} | 955 | 56033 | BARX1_HUMAN | Q9HBU1 |
| BARX2_HUMAN.H11MO.0.D |  |
HUMAN:BARX2 | 10 | D | 0 | hYnWTAATKR | HOCOMOCOv9 | Integrative | 14 | NK-related factors{3.1.2} | BARX{3.1.2.2} | 956 | 8538 | BARX2_HUMAN | Q9UMQ3 | ||||
| BATF3_HUMAN.H11MO.0.B |  |
HUMAN:BATF3 | 17 | B | 0 | ndSTTYChnWhTGAbdn | HOCOMOCOv11 | ChIP-Seq | 0.963 | 5 | 500 | B-ATF-related factors{1.1.4} | B-ATF-3{1.1.4.0.3} | 28915 | 55509 | BATF3_HUMAN | Q9NR55 | ||
| BATF_HUMAN.H11MO.0.A |  |
HUMAN:BATF | 18 | A | 0 | dbTYYYddWWTGASTnWb | HOCOMOCOv11 | ChIP-Seq | 0.963 | 0.938 | 10 | 20 | 500 | B-ATF-related factors{1.1.4} | B-ATF{1.1.4.0.1} | 958 | 10538 | BATF_HUMAN | Q16520 |
| BATF_HUMAN.H11MO.1.A |  |
HUMAN:BATF | 11 | A | 1 | dMTGAYKCAWh | HOCOMOCOv11 | ChIP-Seq | 0.89 | 0.856 | 10 | 20 | 500 | B-ATF-related factors{1.1.4} | B-ATF{1.1.4.0.1} | 958 | 10538 | BATF_HUMAN | Q16520 |
| BC11A_HUMAN.H11MO.0.A |  |
HUMAN:BCL11A | 17 | A | 0 | vdvRvRGGAAvYRRvRv | HOCOMOCOv10 | ChIP-Seq | 0.9 | 10 | 500 | Factors with multiple dispersed zinc fingers{2.3.4} | BCL11{2.3.4.15} | 13221 | 53335 | BC11A_HUMAN | Q9H165 | ||
| BCL6B_HUMAN.H11MO.0.D |  |
HUMAN:BCL6B | 11 | D | 0 | nYGCTTTCTAG | HOCOMOCOv10 | HT-SELEX | 66 | More than 3 adjacent zinc finger factors{2.3.3} | BCL6 factors{2.3.3.22} | 1002 | 255877 | BCL6B_HUMAN | Q8N143 | ||||
| BCL6_HUMAN.H11MO.0.A |  |
HUMAN:BCL6 | 13 | A | 0 | bdMYYTCYAGGAA | HOCOMOCOv11 | ChIP-Seq | 0.802 | 0.874 | 27 | 11 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | BCL6 factors{2.3.3.22} | 1001 | 604 | BCL6_HUMAN | P41182 |
| BHA15_HUMAN.H11MO.0.B |  |
HUMAN:BHLHA15 | 11 | B | 0 | vvRRCAGCTGK | HOCOMOCOv11 | ChIP-Seq | 0.988 | 6 | 500 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 22265 | 168620 | BHA15_HUMAN | Q7RTS1 | ||
| BHE22_HUMAN.H11MO.0.D |  |
HUMAN:BHLHE22 | 13 | D | 0 | dvCATATGTTYhh | HOCOMOCOv10 | HT-SELEX | 2250 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 11963 | 27319 | BHE22_HUMAN | Q8NFJ8 | ||||
| BHE23_HUMAN.H11MO.0.D |  |
HUMAN:BHLHE23 | 12 | D | 0 | dvnRKRTGKKdb | HOCOMOCOv10 | HT-SELEX | 394 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 16093 | 128408 | BHE23_HUMAN | Q8NDY6 | ||||
| BHE40_HUMAN.H11MO.0.A |  |
HUMAN:BHLHE40 | 10 | A | 0 | dKCACGTGMv | HOCOMOCOv11 | ChIP-Seq | 0.855 | 0.953 | 10 | 5 | 508 | Hairy-related factors{1.2.4} | Hairy-like factors{1.2.4.1} | 1046 | 8553 | BHE40_HUMAN | O14503 |
| BHE41_HUMAN.H11MO.0.D |  |
HUMAN:BHLHE41 | 20 | D | 0 | dCnRnSTCRMGTGMhKRnRn | HOCOMOCOv9 | Integrative | 10 | Hairy-related factors{1.2.4} | Hairy-like factors{1.2.4.1} | 16617 | 79365 | BHE41_HUMAN | Q9C0J9 | ||||
| BMAL1_HUMAN.H11MO.0.A |  |
HUMAN:ARNTL | 11 | A | 0 | nSCAYGTGhbb | HOCOMOCOv11 | ChIP-Seq | 0.864 | 0.875 | 18 | 15 | 410 | PAS domain factors{1.2.5} | Arnt-like factors{1.2.5.2} | 701 | 406 | BMAL1_HUMAN | O00327 |
| BPTF_HUMAN.H11MO.0.D |  |
HUMAN:BPTF | 12 | D | 0 | nKKnTTGTdKnv | HOCOMOCOv9 | Integrative | 21 | 3581 | 2186 | BPTF_HUMAN | Q12830 | ||||||
| BRAC_HUMAN.H11MO.0.A | 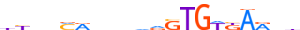 |
HUMAN:T | 20 | A | 0 | bYvvMRdvnvdSTGWKARdd | HOCOMOCOv11 | ChIP-Seq | 0.835 | 0.818 | 9 | 10 | 500 | Brachyury-related factors{6.5.1} | T (Brachyury){6.5.1.0.1} | 11515 | 6862 | BRAC_HUMAN | O15178 |
| BRAC_HUMAN.H11MO.1.B |  |
HUMAN:T | 11 | B | 1 | dRSTGWSARdd | HOCOMOCOv11 | ChIP-Seq | 0.8 | 0.757 | 9 | 10 | 500 | Brachyury-related factors{6.5.1} | T (Brachyury){6.5.1.0.1} | 11515 | 6862 | BRAC_HUMAN | O15178 |
| BRCA1_HUMAN.H11MO.0.D |  |
HUMAN:BRCA1 | 9 | D | 0 | bKKbKGTTG | HOCOMOCOv9 | Integrative | 72 | 1100 | 672 | BRCA1_HUMAN | P38398 | ||||||
| BSH_HUMAN.H11MO.0.D |  |
HUMAN:BSX | 14 | D | 0 | WWWbbvbhTAAYbd | HOCOMOCOv10 | HT-SELEX | 8218 | NK-related factors{3.1.2} | BSX{3.1.2.3} | 20450 | 390259 | BSH_HUMAN | Q3C1V8 | ||||
| CDC5L_HUMAN.H11MO.0.D |  |
HUMAN:CDC5L | 15 | D | 0 | vbGnKdTAAYRWAWb | HOCOMOCOv9 | Integrative | 10 | Myb/SANT domain factors{3.5.1} | Myb-like factors{3.5.1.1} | 1743 | 988 | CDC5L_HUMAN | Q99459 | ||||
| CDX1_HUMAN.H11MO.0.C |  |
HUMAN:CDX1 | 8 | C | 0 | hTTKAWGd | HOCOMOCOv9 | Integrative | 30 | HOX-related factors{3.1.1} | CDX (Caudal type homeobox){3.1.1.9} | 1805 | 1044 | CDX1_HUMAN | P47902 | ||||
| CDX2_HUMAN.H11MO.0.A |  |
HUMAN:CDX2 | 12 | A | 0 | dTTbATKGShbY | HOCOMOCOv11 | ChIP-Seq | 0.923 | 0.982 | 13 | 23 | 151 | HOX-related factors{3.1.1} | CDX (Caudal type homeobox){3.1.1.9} | 1806 | 1045 | CDX2_HUMAN | Q99626 |
| CEBPA_HUMAN.H11MO.0.A |  |
HUMAN:CEBPA | 12 | A | 0 | dRTTKhGCAAYv | HOCOMOCOv11 | ChIP-Seq | 0.983 | 0.991 | 18 | 153 | 500 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 1833 | 1050 | CEBPA_HUMAN | P49715 |
| CEBPB_HUMAN.H11MO.0.A |  |
HUMAN:CEBPB | 12 | A | 0 | dRTTRhGCAAYv | HOCOMOCOv11 | ChIP-Seq | 0.981 | 0.985 | 65 | 158 | 457 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 1834 | 1051 | CEBPB_HUMAN | P17676 |
| CEBPD_HUMAN.H11MO.0.C |  |
HUMAN:CEBPD | 11 | C | 0 | vTTGCdYMAbh | HOCOMOCOv11 | ChIP-Seq | 0.831 | 10 | 500 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 1835 | 1052 | CEBPD_HUMAN | P49716 | ||
| CEBPE_HUMAN.H11MO.0.A |  |
HUMAN:CEBPE | 12 | A | 0 | dTKSGMAAThdb | HOCOMOCOv9 | Integrative | 803 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 1836 | 1053 | CEBPE_HUMAN | Q15744 | ||||
| CEBPG_HUMAN.H11MO.0.B |  |
HUMAN:CEBPG | 12 | B | 0 | RTTRCATCAKhh | HOCOMOCOv11 | ChIP-Seq | 0.738 | 0.972 | 2 | 4 | 501 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 1837 | 1054 | CEBPG_HUMAN | P53567 |
| CEBPZ_HUMAN.H11MO.0.D |  |
HUMAN:CEBPZ | 11 | D | 0 | dSTSATTGGCT | HOCOMOCOv9 | Integrative | 28 | 24218 | 10153 | CEBPZ_HUMAN | Q03701 | ||||||
| CENPB_HUMAN.H11MO.0.D |  |
HUMAN:CENPB | 17 | D | 0 | TTCGYWvnAhRCGGRdh | HOCOMOCOv10 | HT-SELEX | 2219 | 1852 | 1059 | CENPB_HUMAN | P07199 | ||||||
| CLOCK_HUMAN.H11MO.0.C |  |
HUMAN:CLOCK | 14 | C | 0 | SdvnCAMRYGvvvh | HOCOMOCOv11 | ChIP-Seq | 0.658 | 0.735 | 5 | 22 | 253 | PAS domain factors{1.2.5} | Arnt-like factors{1.2.5.2} | 2082 | 9575 | CLOCK_HUMAN | O15516 |
| COE1_HUMAN.H11MO.0.A |  |
HUMAN:EBF1 | 15 | A | 0 | vdbYCCCMWGGGRvh | HOCOMOCOv11 | ChIP-Seq | 0.994 | 0.992 | 17 | 55 | 500 | Early B-Cell Factor-related factors{6.1.5} | EBF1 (COE1){6.1.5.0.1} | 3126 | 1879 | COE1_HUMAN | Q9UH73 |
| COT1_HUMAN.H11MO.0.C |  |
HUMAN:NR2F1 | 17 | C | 0 | vRRRbvRRAGGKCAdvv | HOCOMOCOv11 | ChIP-Seq | 0.879 | 3 | 501 | RXR-related receptors (NR2){2.1.3} | COUP-like receptors (NR2F){2.1.3.5} | 7975 | 7025 | COT1_HUMAN | P10589 | ||
| COT1_HUMAN.H11MO.1.C |  |
HUMAN:NR2F1 | 13 | C | 1 | bvdRAGGKCAvvR | HOCOMOCOv11 | ChIP-Seq | 0.851 | 3 | 478 | RXR-related receptors (NR2){2.1.3} | COUP-like receptors (NR2F){2.1.3.5} | 7975 | 7025 | COT1_HUMAN | P10589 | ||
| COT2_HUMAN.H11MO.0.A |  |
HUMAN:NR2F2 | 13 | A | 0 | vMRAGGTCAdvdd | HOCOMOCOv10 | ChIP-Seq | 0.934 | 0.833 | 28 | 3 | 509 | RXR-related receptors (NR2){2.1.3} | COUP-like receptors (NR2F){2.1.3.5} | 7976 | 7026 | COT2_HUMAN | P24468 |
| COT2_HUMAN.H11MO.1.A |  |
HUMAN:NR2F2 | 17 | A | 1 | dRdvbhWRAGGKCAvvv | HOCOMOCOv11 | ChIP-Seq | 0.915 | 0.732 | 28 | 3 | 503 | RXR-related receptors (NR2){2.1.3} | COUP-like receptors (NR2F){2.1.3.5} | 7976 | 7026 | COT2_HUMAN | P24468 |
| CPEB1_HUMAN.H11MO.0.D | 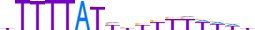 |
HUMAN:CPEB1 | 17 | D | 0 | hTTTTATYhYYYYWWWh | HOCOMOCOv10 | HT-SELEX | 1803 | 21744 | 64506 | CPEB1_HUMAN | Q9BZB8 | ||||||
| CR3L1_HUMAN.H11MO.0.D |  |
HUMAN:CREB3L1 | 11 | D | 0 | KMCACGTGGCA | HOCOMOCOv10 | HT-SELEX | 39883 | CREB-related factors{1.1.7} | CREB-3-like factors{1.1.7.2} | 18856 | 90993 | CR3L1_HUMAN | Q96BA8 | ||||
| CR3L2_HUMAN.H11MO.0.D |  |
HUMAN:CREB3L2 | 13 | D | 0 | YKvWSACGTGdnv | HOCOMOCOv10 | HT-SELEX | 1223 | CREB-related factors{1.1.7} | CREB-3-like factors{1.1.7.2} | 23720 | 64764 | CR3L2_HUMAN | Q70SY1 | ||||
| CREB1_HUMAN.H11MO.0.A |  |
HUMAN:CREB1 | 11 | A | 0 | ndRTGACGYhW | HOCOMOCOv11 | ChIP-Seq | 0.927 | 0.888 | 63 | 8 | 501 | CREB-related factors{1.1.7} | CREB-like factors{1.1.7.1} | 2345 | 1385 | CREB1_HUMAN | P16220 |
| CREB3_HUMAN.H11MO.0.D |  |
HUMAN:CREB3 | 13 | D | 0 | bnRTGACGTGKMd | HOCOMOCOv10 | HT-SELEX | 95 | CREB-related factors{1.1.7} | CREB-3-like factors{1.1.7.2} | 2347 | 10488 | CREB3_HUMAN | O43889 | ||||
| CREB5_HUMAN.H11MO.0.D |  |
HUMAN:CREB5 | 11 | D | 0 | dRTGACGTCAT | HOCOMOCOv10 | HT-SELEX | 1784 | Jun-related factors{1.1.1} | ATF-2-like factors{1.1.1.3} | 16844 | 9586 | CREB5_HUMAN | Q02930 | ||||
| CREM_HUMAN.H11MO.0.C | 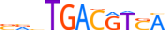 |
HUMAN:CREM | 11 | C | 0 | SRvTGACGTSA | HOCOMOCOv9 | Integrative | 30 | CREB-related factors{1.1.7} | CREB-like factors{1.1.7.1} | 2352 | 1390 | CREM_HUMAN | Q03060 | ||||
| CRX_HUMAN.H11MO.0.B |  |
HUMAN:CRX | 13 | B | 0 | hvRdvGGATTAdv | HOCOMOCOv11 | ChIP-Seq | 0.933 | 8 | 500 | Paired-related HD factors{3.1.3} | OTX{3.1.3.17} | 2383 | 1406 | CRX_HUMAN | O43186 | ||
| CTCFL_HUMAN.H11MO.0.A |  |
HUMAN:CTCFL | 17 | A | 0 | bCCdShAGGKGGCGShv | HOCOMOCOv11 | ChIP-Seq | 0.974 | 0.993 | 25 | 8 | 450 | More than 3 adjacent zinc finger factors{2.3.3} | CTCF-like factors{2.3.3.50} | 16234 | 140690 | CTCFL_HUMAN | Q8NI51 |
| CTCF_HUMAN.H11MO.0.A |  |
HUMAN:CTCF | 19 | A | 0 | bbdCCRSYAGGKGGCRSbv | HOCOMOCOv11 | ChIP-Seq | 0.997 | 0.998 | 502 | 263 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | CTCF-like factors{2.3.3.50} | 13723 | 10664 | CTCF_HUMAN | P49711 |
| CUX1_HUMAN.H11MO.0.C |  |
HUMAN:CUX1 | 14 | C | 0 | vbRvndATYRRTbb | HOCOMOCOv9 | Integrative | 97 | HD-CUT factors{3.1.9} | CUX{3.1.9.2} | 2557 | 1523 | CUX1_HUMAN | P39880 | ||||
| CUX2_HUMAN.H11MO.0.D |  |
HUMAN:CUX2 | 11 | D | 0 | hRRTCAATvvv | HOCOMOCOv11 | ChIP-Seq | 0.867 | 5 | 496 | HD-CUT factors{3.1.9} | CUX{3.1.9.2} | 19347 | 23316 | CUX2_HUMAN | O14529 | ||
| CXXC1_HUMAN.H11MO.0.D |  |
HUMAN:CXXC1 | 7 | D | 0 | YGKTKKY | HOCOMOCOv9 | Integrative | 63 | CpG-binding proteins{2.6.1} | CpG-binding protein (CXXC1, CFP1, CGBP, PCCX1, PHF18){2.6.1.0.1} | 24343 | 30827 | CXXC1_HUMAN | Q9P0U4 | ||||
| DBP_HUMAN.H11MO.0.B |  |
HUMAN:DBP | 11 | B | 0 | vTKRYGTAAbv | HOCOMOCOv9 | Integrative | 26 | C/EBP-related{1.1.8} | PAR factors{1.1.8.2} | 2697 | 1628 | DBP_HUMAN | Q10586 | ||||
| DDIT3_HUMAN.H11MO.0.D |  |
HUMAN:DDIT3 | 10 | D | 0 | MTGATGMAAb | HOCOMOCOv11 | ChIP-Seq | 0.628 | 0.91 | 3 | 6 | 500 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 2726 | 1649 | DDIT3_HUMAN | P35638 |
| DLX1_HUMAN.H11MO.0.D |  |
HUMAN:DLX1 | 16 | D | 0 | dhWWWWddbbWAATTd | HOCOMOCOv10 | HT-SELEX | 16332 | NK-related factors{3.1.2} | DLX{3.1.2.5} | 2914 | 1745 | DLX1_HUMAN | P56177 | ||||
| DLX2_HUMAN.H11MO.0.D |  |
HUMAN:DLX2 | 8 | D | 0 | ATAATTAn | HOCOMOCOv9 | Integrative | 6 | NK-related factors{3.1.2} | DLX{3.1.2.5} | 2915 | 1746 | DLX2_HUMAN | Q07687 | ||||
| DLX3_HUMAN.H11MO.0.C |  |
HUMAN:DLX3 | 10 | C | 0 | RMTAATTRvh | HOCOMOCOv9 | Integrative | 35 | NK-related factors{3.1.2} | DLX{3.1.2.5} | 2916 | 1747 | DLX3_HUMAN | O60479 | ||||
| DLX4_HUMAN.H11MO.0.D |  |
HUMAN:DLX4 | 16 | D | 0 | YAATTAbnnhhWWWWd | HOCOMOCOv10 | HT-SELEX | 2360 | NK-related factors{3.1.2} | DLX{3.1.2.5} | 2917 | 1748 | DLX4_HUMAN | Q92988 | ||||
| DLX5_HUMAN.H11MO.0.D |  |
HUMAN:DLX5 | 11 | D | 0 | WRATTRYWKbb | HOCOMOCOv11 | ChIP-Seq | 0.909 | 2 | 465 | NK-related factors{3.1.2} | DLX{3.1.2.5} | 2918 | 1749 | DLX5_HUMAN | P56178 | ||
| DLX6_HUMAN.H11MO.0.D |  |
HUMAN:DLX6 | 15 | D | 0 | hWddddnnnTAATTR | HOCOMOCOv10 | HT-SELEX | 1138 | NK-related factors{3.1.2} | DLX{3.1.2.5} | 2919 | 1750 | DLX6_HUMAN | P56179 | ||||
| DMBX1_HUMAN.H11MO.0.D |  |
HUMAN:DMBX1 | 20 | D | 0 | hhvhWWWWWbYYRATTAvnn | HOCOMOCOv10 | HT-SELEX | 29 | Paired-related HD factors{3.1.3} | DMBX{3.1.3.4} | 19026 | 127343 | DMBX1_HUMAN | Q8NFW5 | ||||
| DMRT1_HUMAN.H11MO.0.D |  |
HUMAN:DMRT1 | 17 | D | 0 | hdGhhACAAWGTWdChv | HOCOMOCOv11 | ChIP-Seq | 0.552 | 0.987 | 2 | 4 | 500 | DMRT{2.5.1} | DMRT1{2.5.1.0.1} | 2934 | 1761 | DMRT1_HUMAN | Q9Y5R6 |
| DPRX_HUMAN.H11MO.0.D |  |
HUMAN:DPRX | 11 | D | 0 | nvGGATTAdSn | HOCOMOCOv10 | HT-SELEX | 46619 | Paired-related HD factors{3.1.3} | DPRX{3.1.3.5} | 32166 | 503834 | DPRX_HUMAN | A6NFQ7 | ||||
| DRGX_HUMAN.H11MO.0.D |  |
HUMAN:DRGX | 11 | D | 0 | TAATYTAATTA | HOCOMOCOv10 | HT-SELEX | 10544 | Paired-related HD factors{3.1.3} | DRGX{3.1.3.6} | 21536 | 644168 | DRGX_HUMAN | A6NNA5 | ||||
| DUX4_HUMAN.H11MO.0.A |  |
HUMAN:DUX4 | 11 | A | 0 | TGATTRRRTTA | HOCOMOCOv11 | ChIP-Seq | 0.996 | 5 | 500 | Paired-related HD factors{3.1.3} | DUX{3.1.3.7} | 50800 | 100288687 | DUX4_HUMAN | Q9UBX2 | ||
| DUXA_HUMAN.H11MO.0.D |  |
HUMAN:DUXA | 11 | D | 0 | TGATTARvTYR | HOCOMOCOv10 | HT-SELEX | 2685 | Paired-related HD factors{3.1.3} | DUX{3.1.3.7} | 32179 | 503835 | DUXA_HUMAN | A6NLW8 | ||||
| E2F1_HUMAN.H11MO.0.A |  |
HUMAN:E2F1 | 14 | A | 0 | vvndSGCGGGARvd | HOCOMOCOv11 | ChIP-Seq | 0.919 | 0.958 | 80 | 21 | 500 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 3113 | 1869 | E2F1_HUMAN | Q01094 |
| E2F2_HUMAN.H11MO.0.B |  |
HUMAN:E2F2 | 10 | B | 0 | RGCGCGMAWC | HOCOMOCOv9 | Integrative | 8 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 3114 | 1870 | E2F2_HUMAN | Q14209 | ||||
| E2F3_HUMAN.H11MO.0.A |  |
HUMAN:E2F3 | 11 | A | 0 | ddRGMKGGARv | HOCOMOCOv11 | ChIP-Seq | 0.888 | 0.916 | 3 | 22 | 500 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 3115 | 1871 | E2F3_HUMAN | O00716 |
| E2F4_HUMAN.H11MO.0.A |  |
HUMAN:E2F4 | 13 | A | 0 | vddSGCGGGRvvd | HOCOMOCOv11 | ChIP-Seq | 0.956 | 0.977 | 15 | 37 | 500 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 3118 | 1874 | E2F4_HUMAN | Q16254 |
| E2F4_HUMAN.H11MO.1.A |  |
HUMAN:E2F4 | 14 | A | 1 | SGCGGGRRddbbnd | HOCOMOCOv11 | ChIP-Seq | 0.924 | 0.97 | 15 | 37 | 500 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 3118 | 1874 | E2F4_HUMAN | Q16254 |
| E2F5_HUMAN.H11MO.0.B |  |
HUMAN:E2F5 | 10 | B | 0 | nGCGCCAAAh | HOCOMOCOv9 | Integrative | 12 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 3119 | 1875 | E2F5_HUMAN | Q15329 | ||||
| E2F6_HUMAN.H11MO.0.A |  |
HUMAN:E2F6 | 13 | A | 0 | dRRGMGGGARvvv | HOCOMOCOv11 | ChIP-Seq | 0.902 | 17 | 500 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 3120 | 1876 | E2F6_HUMAN | O75461 | ||
| E2F7_HUMAN.H11MO.0.B |  |
HUMAN:E2F7 | 13 | B | 0 | vdRGCGGGARvdv | HOCOMOCOv11 | ChIP-Seq | 0.846 | 16 | 500 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 23820 | 144455 | E2F7_HUMAN | Q96AV8 | ||
| E2F8_HUMAN.H11MO.0.D | 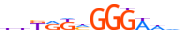 |
HUMAN:E2F8 | 12 | D | 0 | dnKRKMGGGWWd | HOCOMOCOv10 | HT-SELEX | 28 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 24727 | 79733 | E2F8_HUMAN | A0AVK6 | ||||
| E4F1_HUMAN.H11MO.0.D |  |
HUMAN:E4F1 | 9 | D | 0 | nGTGACGTM | HOCOMOCOv9 | Integrative | 9 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 3121 | 1877 | E4F1_HUMAN | Q66K89 | ||||
| EGR1_HUMAN.H11MO.0.A |  |
HUMAN:EGR1 | 17 | A | 0 | vnvdGhGKGGGYGKndv | HOCOMOCOv11 | ChIP-Seq | 0.989 | 0.981 | 36 | 12 | 500 | Three-zinc finger Krüppel-related factors{2.3.1} | EGR factors{2.3.1.3} | 3238 | 1958 | EGR1_HUMAN | P18146 |
| EGR2_HUMAN.H11MO.0.A |  |
HUMAN:EGR2 | 18 | A | 0 | RdRdvdShGKGGGhGKdR | HOCOMOCOv11 | ChIP-Seq | 0.987 | 0.963 | 4 | 25 | 507 | Three-zinc finger Krüppel-related factors{2.3.1} | EGR factors{2.3.1.3} | 3239 | 1959 | EGR2_HUMAN | P11161 |
| EGR2_HUMAN.H11MO.1.A |  |
HUMAN:EGR2 | 16 | A | 1 | dvdGhGKGGGhGKddv | HOCOMOCOv11 | ChIP-Seq | 0.988 | 0.98 | 4 | 25 | 505 | Three-zinc finger Krüppel-related factors{2.3.1} | EGR factors{2.3.1.3} | 3239 | 1959 | EGR2_HUMAN | P11161 |
| EGR3_HUMAN.H11MO.0.D |  |
HUMAN:EGR3 | 11 | D | 0 | WRMGTGGGTGb | HOCOMOCOv9 | Integrative | 8 | Three-zinc finger Krüppel-related factors{2.3.1} | EGR factors{2.3.1.3} | 3240 | 1960 | EGR3_HUMAN | Q06889 | ||||
| EGR4_HUMAN.H11MO.0.D |  |
HUMAN:EGR4 | 11 | D | 0 | GGSGGYAGGGM | HOCOMOCOv9 | Integrative | 6 | Three-zinc finger Krüppel-related factors{2.3.1} | EGR factors{2.3.1.3} | 3241 | 1961 | EGR4_HUMAN | Q05215 | ||||
| EHF_HUMAN.H11MO.0.B |  |
HUMAN:EHF | 15 | B | 0 | ndAbSAGGAAGYdvv | HOCOMOCOv11 | ChIP-Seq | 0.966 | 0.654 | 5 | 3 | 500 | Ets-related factors{3.5.2} | EHF-like factors{3.5.2.4} | 3246 | 26298 | EHF_HUMAN | Q9NZC4 |
| ELF1_HUMAN.H11MO.0.A |  |
HUMAN:ELF1 | 14 | A | 0 | vvdvSMGGAAGYvv | HOCOMOCOv11 | ChIP-Seq | 0.992 | 0.975 | 43 | 5 | 500 | Ets-related factors{3.5.2} | Elf-1-like factors{3.5.2.3} | 3316 | 1997 | ELF1_HUMAN | P32519 |
| ELF2_HUMAN.H11MO.0.C |  |
HUMAN:ELF2 | 13 | C | 0 | vdvSCGGAAGhvv | HOCOMOCOv11 | ChIP-Seq | 0.893 | 2 | 500 | Ets-related factors{3.5.2} | Elf-1-like factors{3.5.2.3} | 3317 | 1998 | ELF2_HUMAN | Q15723 | ||
| ELF3_HUMAN.H11MO.0.A |  |
HUMAN:ELF3 | 14 | A | 0 | vdAbSAGGAARYdv | HOCOMOCOv11 | ChIP-Seq | 0.979 | 11 | 500 | Ets-related factors{3.5.2} | EHF-like factors{3.5.2.4} | 3318 | 1999 | ELF3_HUMAN | P78545 | ||
| ELF5_HUMAN.H11MO.0.A |  |
HUMAN:ELF5 | 15 | A | 0 | vvvAbSMGGAAGhdv | HOCOMOCOv11 | ChIP-Seq | 0.985 | 0.93 | 3 | 12 | 500 | Ets-related factors{3.5.2} | EHF-like factors{3.5.2.4} | 3320 | 2001 | ELF5_HUMAN | Q9UKW6 |
| ELK1_HUMAN.H11MO.0.B |  |
HUMAN:ELK1 | 11 | B | 0 | vCCGGAAGTKv | HOCOMOCOv11 | ChIP-Seq | 0.98 | 0.683 | 4 | 7 | 500 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 3321 | 2002 | ELK1_HUMAN | P19419 |
| ELK3_HUMAN.H11MO.0.D |  |
HUMAN:ELK3 | 12 | D | 0 | vnCMGGAWGTGC | HOCOMOCOv9 | Integrative | 7 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 3325 | 2004 | ELK3_HUMAN | P41970 | ||||
| ELK4_HUMAN.H11MO.0.A |  |
HUMAN:ELK4 | 12 | A | 0 | vvCCGGAARYRv | HOCOMOCOv11 | ChIP-Seq | 0.966 | 0.638 | 13 | 4 | 500 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 3326 | 2005 | ELK4_HUMAN | P28324 |
| EMX1_HUMAN.H11MO.0.D |  |
HUMAN:EMX1 | 12 | D | 0 | TAATTARbbhWh | HOCOMOCOv10 | HT-SELEX | 31003 | NK-related factors{3.1.2} | EMX{3.1.2.6} | 3340 | 2016 | EMX1_HUMAN | Q04741 | ||||
| EMX2_HUMAN.H11MO.0.D |  |
HUMAN:EMX2 | 12 | D | 0 | TAATTARbdhhh | HOCOMOCOv10 | HT-SELEX | 36122 | NK-related factors{3.1.2} | EMX{3.1.2.6} | 3341 | 2018 | EMX2_HUMAN | Q04743 | ||||
| EOMES_HUMAN.H11MO.0.D |  |
HUMAN:EOMES | 12 | D | 0 | ddvARGTGhbAR | HOCOMOCOv10 | ChIP-Seq | 376 | TBrain-related factors{6.5.2} | TBR-2 (EOMES){6.5.2.0.2} | 3372 | 8320 | EOMES_HUMAN | O95936 | ||||
| EPAS1_HUMAN.H11MO.0.B |  |
HUMAN:EPAS1 | 9 | B | 0 | vdACGTGhh | HOCOMOCOv11 | ChIP-Seq | 0.838 | 24 | 500 | PAS domain factors{1.2.5} | Ahr-like factors{1.2.5.1} | 3374 | 2034 | EPAS1_HUMAN | Q99814 | ||
| ERG_HUMAN.H11MO.0.A |  |
HUMAN:ERG | 13 | A | 0 | vvRSAGGAAGbvv | HOCOMOCOv11 | ChIP-Seq | 0.984 | 0.877 | 79 | 16 | 500 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 3446 | 2078 | ERG_HUMAN | P11308 |
| ERR1_HUMAN.H11MO.0.A |  |
HUMAN:ESRRA | 15 | A | 0 | bYSAAGGTCAhhndv | HOCOMOCOv11 | ChIP-Seq | 0.956 | 0.974 | 31 | 3 | 284 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 3471 | 2101 | ERR1_HUMAN | P11474 |
| ERR2_HUMAN.H11MO.0.A |  |
HUMAN:ESRRB | 9 | A | 0 | bSAAGGTCA | HOCOMOCOv9 | Integrative | 3687 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 3473 | 2103 | ERR2_HUMAN | O95718 | ||||
| ERR3_HUMAN.H11MO.0.B |  |
HUMAN:ESRRG | 9 | B | 0 | TYAAGGTCA | HOCOMOCOv9 | Integrative | 14 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 3474 | 2104 | ERR3_HUMAN | P62508 | ||||
| ESR1_HUMAN.H11MO.0.A |  |
HUMAN:ESR1 | 15 | A | 0 | RGGKCAbCvWGMCCY | HOCOMOCOv11 | ChIP-Seq | 0.971 | 0.995 | 432 | 58 | 500 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 3467 | 2099 | ESR1_HUMAN | P03372 |
| ESR1_HUMAN.H11MO.1.A |  |
HUMAN:ESR1 | 10 | A | 1 | hhRGGTCAbv | HOCOMOCOv11 | ChIP-Seq | 0.935 | 0.957 | 432 | 58 | 500 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 3467 | 2099 | ESR1_HUMAN | P03372 |
| ESR2_HUMAN.H11MO.0.A |  |
HUMAN:ESR2 | 15 | A | 0 | RGGbCRbCbYGhCCY | HOCOMOCOv11 | ChIP-Seq | 0.944 | 0.941 | 11 | 7 | 497 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 3468 | 2100 | ESR2_HUMAN | Q92731 |
| ESR2_HUMAN.H11MO.1.A |  |
HUMAN:ESR2 | 7 | A | 1 | RGGTCAS | HOCOMOCOv11 | ChIP-Seq | 0.879 | 0.851 | 11 | 7 | 500 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 3468 | 2100 | ESR2_HUMAN | Q92731 |
| ESX1_HUMAN.H11MO.0.D |  |
HUMAN:ESX1 | 17 | D | 0 | vbTAATTAvbnhWWWWd | HOCOMOCOv10 | HT-SELEX | 8289 | Paired-related HD factors{3.1.3} | ESX{3.1.3.8} | 14865 | 80712 | ESX1_HUMAN | Q8N693 | ||||
| ETS1_HUMAN.H11MO.0.A |  |
HUMAN:ETS1 | 13 | A | 0 | vvRSMGGAAGYRR | HOCOMOCOv11 | ChIP-Seq | 0.963 | 0.927 | 45 | 33 | 500 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 3488 | 2113 | ETS1_HUMAN | P14921 |
| ETS2_HUMAN.H11MO.0.B |  |
HUMAN:ETS2 | 13 | B | 0 | ddvdRGAARvRdv | HOCOMOCOv11 | ChIP-Seq | 0.836 | 12 | 500 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 3489 | 2114 | ETS2_HUMAN | P15036 | ||
| ETV1_HUMAN.H11MO.0.A |  |
HUMAN:ETV1 | 11 | A | 0 | vvCCGGAAGbv | HOCOMOCOv11 | ChIP-Seq | 0.941 | 8 | 500 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 3490 | 2115 | ETV1_HUMAN | P50549 | ||
| ETV2_HUMAN.H11MO.0.B |  |
HUMAN:ETV2 | 16 | B | 0 | vvvRRCAGGAARYvSv | HOCOMOCOv11 | ChIP-Seq | 0.981 | 6 | 230 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 3491 | 2116 | ETV2_HUMAN | O00321 | ||
| ETV3_HUMAN.H11MO.0.D |  |
HUMAN:ETV3 | 14 | D | 0 | dnhdTdTGCACYTY | HOCOMOCOv10 | HT-SELEX | 12 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 3492 | 2117 | ETV3_HUMAN | P41162 | ||||
| ETV4_HUMAN.H11MO.0.B |  |
HUMAN:ETV4 | 11 | B | 0 | vdSAGGAARbd | HOCOMOCOv11 | ChIP-Seq | 0.819 | 5 | 500 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 3493 | 2118 | ETV4_HUMAN | P43268 | ||
| ETV5_HUMAN.H11MO.0.C |  |
HUMAN:ETV5 | 14 | C | 0 | vddvAGGRARddvv | HOCOMOCOv11 | ChIP-Seq | 0.867 | 3 | 500 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 3494 | 2119 | ETV5_HUMAN | P41161 | ||
| ETV6_HUMAN.H11MO.0.D |  |
HUMAN:ETV6 | 9 | D | 0 | dSAGGAARb | HOCOMOCOv11 | ChIP-Seq | 0.853 | 3 | 500 | Ets-related factors{3.5.2} | ETV6-like factors{3.5.2.6} | 3495 | 2120 | ETV6_HUMAN | P41212 | ||
| ETV7_HUMAN.H11MO.0.D |  |
HUMAN:ETV7 | 7 | D | 0 | vRGGAAR | HOCOMOCOv11 | ChIP-Seq | 0.729 | 3 | 500 | Ets-related factors{3.5.2} | ETV6-like factors{3.5.2.6} | 18160 | 51513 | ETV7_HUMAN | Q9Y603 | ||
| EVI1_HUMAN.H11MO.0.B |  |
HUMAN:MECOM | 16 | B | 0 | dWGAYAAGATAAnMbd | HOCOMOCOv9 | Integrative | 141 | Factors with multiple dispersed zinc fingers{2.3.4} | Evi-1-like factors{2.3.4.14} | 3498 | 2122 | EVI1_HUMAN | Q03112 | ||||
| FEV_HUMAN.H11MO.0.B |  |
HUMAN:FEV | 10 | B | 0 | vvvRGARRhS | HOCOMOCOv11 | ChIP-Seq | 0.753 | 0.887 | 2 | 4 | 498 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 18562 | 54738 | FEV_HUMAN | Q99581 |
| FEZF1_HUMAN.H11MO.0.C |  |
HUMAN:FEZF1 | 12 | C | 0 | dhTGYYYWTTYh | HOCOMOCOv11 | ChIP-Seq | 0.801 | 3 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | FEZF factors{2.3.3.20} | 22788 | 389549 | FEZF1_HUMAN | A0PJY2 | ||
| FIGLA_HUMAN.H11MO.0.D |  |
HUMAN:FIGLA | 11 | D | 0 | dnhCAGGTGbd | HOCOMOCOv10 | HT-SELEX | 159 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 24669 | 344018 | FIGLA_HUMAN | Q6QHK4 | ||||
| FLI1_HUMAN.H11MO.0.A |  |
HUMAN:FLI1 | 18 | A | 0 | vvdRRMAGGAAGbvRvvv | HOCOMOCOv11 | ChIP-Seq | 0.987 | 0.892 | 50 | 32 | 529 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 3749 | 2313 | FLI1_HUMAN | Q01543 |
| FLI1_HUMAN.H11MO.1.A |  |
HUMAN:FLI1 | 13 | A | 1 | vdRSAGGAAGbvd | HOCOMOCOv11 | ChIP-Seq | 0.986 | 0.953 | 50 | 32 | 500 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 3749 | 2313 | FLI1_HUMAN | Q01543 |
| FOSB_HUMAN.H11MO.0.A |  |
HUMAN:FOSB | 9 | A | 0 | vTGAGTCAb | HOCOMOCOv11 | ChIP-Seq | 0.974 | 0.917 | 2 | 27 | 500 | Fos-related factors{1.1.2} | Fos factors{1.1.2.1} | 3797 | 2354 | FOSB_HUMAN | P53539 |
| FOSL1_HUMAN.H11MO.0.A |  |
HUMAN:FOSL1 | 12 | A | 0 | ddvTGAGTCAbh | HOCOMOCOv11 | ChIP-Seq | 0.988 | 0.952 | 21 | 2 | 500 | Fos-related factors{1.1.2} | Fos factors{1.1.2.1} | 13718 | 8061 | FOSL1_HUMAN | P15407 |
| FOSL2_HUMAN.H11MO.0.A |  |
HUMAN:FOSL2 | 12 | A | 0 | ndRTGAGTCAYh | HOCOMOCOv11 | ChIP-Seq | 0.997 | 0.976 | 27 | 5 | 500 | Fos-related factors{1.1.2} | Fos factors{1.1.2.1} | 3798 | 2355 | FOSL2_HUMAN | P15408 |
| FOS_HUMAN.H11MO.0.A |  |
HUMAN:FOS | 9 | A | 0 | vTGAGTCAb | HOCOMOCOv11 | ChIP-Seq | 0.993 | 0.929 | 38 | 50 | 500 | Fos-related factors{1.1.2} | Fos factors{1.1.2.1} | 3796 | 2353 | FOS_HUMAN | P01100 |
| FOXA1_HUMAN.H11MO.0.A |  |
HUMAN:FOXA1 | 12 | A | 0 | TGTTTACWYWdb | HOCOMOCOv11 | ChIP-Seq | 0.988 | 0.974 | 199 | 28 | 500 | Forkhead box (FOX) factors{3.3.1} | FOXA{3.3.1.1} | 5021 | 3169 | FOXA1_HUMAN | P55317 |
| FOXA2_HUMAN.H11MO.0.A |  |
HUMAN:FOXA2 | 12 | A | 0 | TGTTTACWbWdb | HOCOMOCOv11 | ChIP-Seq | 0.982 | 0.988 | 22 | 34 | 500 | Forkhead box (FOX) factors{3.3.1} | FOXA{3.3.1.1} | 5022 | 3170 | FOXA2_HUMAN | Q9Y261 |
| FOXA3_HUMAN.H11MO.0.B |  |
HUMAN:FOXA3 | 13 | B | 0 | bhTGTTTACWbWd | HOCOMOCOv11 | ChIP-Seq | 0.988 | 6 | 500 | Forkhead box (FOX) factors{3.3.1} | FOXA{3.3.1.1} | 5023 | 3171 | FOXA3_HUMAN | P55318 | ||
| FOXB1_HUMAN.H11MO.0.D |  |
HUMAN:FOXB1 | 17 | D | 0 | dbvWWTATTTACATWRb | HOCOMOCOv10 | HT-SELEX | 6619 | Forkhead box (FOX) factors{3.3.1} | FOXB{3.3.1.2} | 3799 | 27023 | FOXB1_HUMAN | Q99853 | ||||
| FOXC1_HUMAN.H11MO.0.C |  |
HUMAN:FOXC1 | 15 | C | 0 | bnhTGTTTACWTAvS | HOCOMOCOv9 | Integrative | 26 | Forkhead box (FOX) factors{3.3.1} | FOXC{3.3.1.3} | 3800 | 2296 | FOXC1_HUMAN | Q12948 | ||||
| FOXC2_HUMAN.H11MO.0.D |  |
HUMAN:FOXC2 | 15 | D | 0 | KhTTGTTdTGYCAGd | HOCOMOCOv9 | Integrative | 5 | Forkhead box (FOX) factors{3.3.1} | FOXC{3.3.1.3} | 3801 | 2303 | FOXC2_HUMAN | Q99958 | ||||
| FOXD1_HUMAN.H11MO.0.D |  |
HUMAN:FOXD1 | 15 | D | 0 | nhhTSTTTAChWhvR | HOCOMOCOv9 | Integrative | 22 | Forkhead box (FOX) factors{3.3.1} | FOXD{3.3.1.4} | 3802 | 2297 | FOXD1_HUMAN | Q16676 | ||||
| FOXD2_HUMAN.H11MO.0.D |  |
HUMAN:FOXD2 | 19 | D | 0 | WRdRTMAATRTTWWYdTAR | HOCOMOCOv10 | HT-SELEX | 968 | Forkhead box (FOX) factors{3.3.1} | FOXD{3.3.1.4} | 3803 | 2306 | FOXD2_HUMAN | O60548 | ||||
| FOXD3_HUMAN.H11MO.0.D | 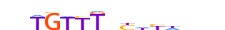 |
HUMAN:FOXD3 | 15 | D | 0 | bbTGTTTnYhYddbh | HOCOMOCOv11 | ChIP-Seq | 0.786 | 11 | 465 | Forkhead box (FOX) factors{3.3.1} | FOXD{3.3.1.4} | 3804 | 27022 | FOXD3_HUMAN | Q9UJU5 | ||
| FOXF1_HUMAN.H11MO.0.D |  |
HUMAN:FOXF1 | 11 | D | 0 | nTGTTTATbWW | HOCOMOCOv9 | Integrative | 8 | Forkhead box (FOX) factors{3.3.1} | FOXF{3.3.1.6} | 3809 | 2294 | FOXF1_HUMAN | Q12946 | ||||
| FOXF2_HUMAN.H11MO.0.D |  |
HUMAN:FOXF2 | 11 | D | 0 | TGTTTAChTWd | HOCOMOCOv9 | Integrative | 26 | Forkhead box (FOX) factors{3.3.1} | FOXF{3.3.1.6} | 3810 | 2295 | FOXF2_HUMAN | Q12947 | ||||
| FOXG1_HUMAN.H11MO.0.D |  |
HUMAN:FOXG1 | 17 | D | 0 | dWTGTTTMYdWdTdKWb | HOCOMOCOv10 | HT-SELEX | 306 | Forkhead box (FOX) factors{3.3.1} | FOXG{3.3.1.7} | 3811 | 2290 | FOXG1_HUMAN | P55316 | ||||
| FOXH1_HUMAN.H11MO.0.A |  |
HUMAN:FOXH1 | 9 | A | 0 | WGTGKATTb | HOCOMOCOv11 | ChIP-Seq | 0.973 | 7 | 487 | Forkhead box (FOX) factors{3.3.1} | FOXH{3.3.1.8} | 3814 | 8928 | FOXH1_HUMAN | O75593 | ||
| FOXI1_HUMAN.H11MO.0.B |  |
HUMAN:FOXI1 | 12 | B | 0 | hWbTGATTGGYb | HOCOMOCOv9 | Integrative | 210 | Forkhead box (FOX) factors{3.3.1} | FOXI{3.3.1.9} | 3815 | 2299 | FOXI1_HUMAN | Q12951 | ||||
| FOXJ2_HUMAN.H11MO.0.C |  |
HUMAN:FOXJ2 | 10 | C | 0 | TRTTTATYTd | HOCOMOCOv9 | Integrative | 41 | Forkhead box (FOX) factors{3.3.1} | FOXJ{3.3.1.10} | 24818 | 55810 | FOXJ2_HUMAN | Q9P0K8 | ||||
| FOXJ3_HUMAN.H11MO.0.A |  |
HUMAN:FOXJ3 | 13 | A | 0 | dTGTTTATKKTTd | HOCOMOCOv9 | Integrative | 550 | Forkhead box (FOX) factors{3.3.1} | FOXJ{3.3.1.10} | 29178 | 22887 | FOXJ3_HUMAN | Q9UPW0 | ||||
| FOXJ3_HUMAN.H11MO.1.B |  |
HUMAN:FOXJ3 | 13 | B | 1 | WKKTTWWTGTTTW | HOCOMOCOv9 | Integrative | 499 | Forkhead box (FOX) factors{3.3.1} | FOXJ{3.3.1.10} | 29178 | 22887 | FOXJ3_HUMAN | Q9UPW0 | ||||
| FOXK1_HUMAN.H11MO.0.A | 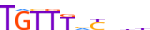 |
HUMAN:FOXK1 | 10 | A | 0 | TGTTThYhhb | HOCOMOCOv11 | ChIP-Seq | 0.857 | 0.781 | 11 | 4 | 506 | Forkhead box (FOX) factors{3.3.1} | FOXK{3.3.1.11} | 23480 | 221937 | FOXK1_HUMAN | P85037 |
| FOXL1_HUMAN.H11MO.0.D |  |
HUMAN:FOXL1 | 19 | D | 0 | bbWTTGTTTGTTTAYWTTW | HOCOMOCOv10 | HT-SELEX | 1207 | Forkhead box (FOX) factors{3.3.1} | FOXL{3.3.1.12} | 3817 | 2300 | FOXL1_HUMAN | Q12952 | ||||
| FOXM1_HUMAN.H11MO.0.A |  |
HUMAN:FOXM1 | 12 | A | 0 | TRTTTRYWbWbn | HOCOMOCOv11 | ChIP-Seq | 0.882 | 66 | 500 | Forkhead box (FOX) factors{3.3.1} | FOXM{3.3.1.13} | 3818 | 2305 | FOXM1_HUMAN | Q08050 | ||
| FOXO1_HUMAN.H11MO.0.A |  |
HUMAN:FOXO1 | 12 | A | 0 | dbbbTGTTKhYn | HOCOMOCOv11 | ChIP-Seq | 0.883 | 0.901 | 9 | 21 | 500 | Forkhead box (FOX) factors{3.3.1} | FOXO{3.3.1.15} | 3819 | 2308 | FOXO1_HUMAN | Q12778 |
| FOXO3_HUMAN.H11MO.0.B |  |
HUMAN:FOXO3 | 10 | B | 0 | bbTGTTTWCh | HOCOMOCOv11 | ChIP-Seq | 0.668 | 0.947 | 2 | 7 | 500 | Forkhead box (FOX) factors{3.3.1} | FOXO{3.3.1.15} | 3821 | 2309 | FOXO3_HUMAN | O43524 |
| FOXO4_HUMAN.H11MO.0.C |  |
HUMAN:FOXO4 | 9 | C | 0 | bTGTTTAbK | HOCOMOCOv9 | Integrative | 28 | Forkhead box (FOX) factors{3.3.1} | FOXO{3.3.1.15} | 7139 | 4303 | FOXO4_HUMAN | P98177 | ||||
| FOXO6_HUMAN.H11MO.0.D |  |
HUMAN:FOXO6 | 13 | D | 0 | dhbWTGTTTACdh | HOCOMOCOv10 | HT-SELEX | 3391 | Forkhead box (FOX) factors{3.3.1} | FOXO{3.3.1.15} | 24814 | FOXO6_HUMAN | A8MYZ6 | |||||
| FOXP1_HUMAN.H11MO.0.A |  |
HUMAN:FOXP1 | 9 | A | 0 | KbTGTTTMY | HOCOMOCOv11 | ChIP-Seq | 0.887 | 21 | 296 | Forkhead box (FOX) factors{3.3.1} | FOXP{3.3.1.16} | 3823 | 27086 | FOXP1_HUMAN | Q9H334 | ||
| FOXP2_HUMAN.H11MO.0.C |  |
HUMAN:FOXP2 | 9 | C | 0 | bTGTTTMCh | HOCOMOCOv11 | ChIP-Seq | 0.962 | 2 | 500 | Forkhead box (FOX) factors{3.3.1} | FOXP{3.3.1.16} | 13875 | 93986 | FOXP2_HUMAN | O15409 | ||
| FOXP3_HUMAN.H11MO.0.D |  |
HUMAN:FOXP3 | 9 | D | 0 | WATKTGWTT | HOCOMOCOv9 | Integrative | 9 | Forkhead box (FOX) factors{3.3.1} | FOXP{3.3.1.16} | 6106 | 50943 | FOXP3_HUMAN | Q9BZS1 | ||||
| FOXQ1_HUMAN.H11MO.0.C |  |
HUMAN:FOXQ1 | 12 | C | 0 | nRTTGTTTATKT | HOCOMOCOv9 | Integrative | 13 | Forkhead box (FOX) factors{3.3.1} | FOXQ{3.3.1.17} | 20951 | 94234 | FOXQ1_HUMAN | Q9C009 | ||||
| FUBP1_HUMAN.H11MO.0.D |  |
HUMAN:FUBP1 | 12 | D | 0 | TYGTnhKTbKKK | HOCOMOCOv9 | Integrative | 102 | 4004 | 8880 | FUBP1_HUMAN | Q96AE4 | ||||||
| GABPA_HUMAN.H11MO.0.A |  |
HUMAN:GABPA | 14 | A | 0 | vvnvSCGGAAGYvv | HOCOMOCOv11 | ChIP-Seq | 0.988 | 0.949 | 92 | 8 | 500 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 4071 | 2551 | GABPA_HUMAN | Q06546 |
| GATA1_HUMAN.H11MO.0.A |  |
HUMAN:GATA1 | 19 | A | 0 | bbKbnnnnnnvWGATAAvv | HOCOMOCOv11 | ChIP-Seq | 0.981 | 0.943 | 48 | 122 | 500 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 4170 | 2623 | GATA1_HUMAN | P15976 |
| GATA1_HUMAN.H11MO.1.A |  |
HUMAN:GATA1 | 11 | A | 1 | dvWGATAASvv | HOCOMOCOv11 | ChIP-Seq | 0.983 | 0.946 | 48 | 122 | 500 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 4170 | 2623 | GATA1_HUMAN | P15976 |
| GATA2_HUMAN.H11MO.0.A |  |
HUMAN:GATA2 | 19 | A | 0 | bKKbnnnnndvAGATAASv | HOCOMOCOv11 | ChIP-Seq | 0.958 | 0.971 | 84 | 54 | 500 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 4171 | 2624 | GATA2_HUMAN | P23769 |
| GATA2_HUMAN.H11MO.1.A |  |
HUMAN:GATA2 | 11 | A | 1 | dvAGATAASvd | HOCOMOCOv11 | ChIP-Seq | 0.946 | 0.963 | 84 | 54 | 500 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 4171 | 2624 | GATA2_HUMAN | P23769 |
| GATA3_HUMAN.H11MO.0.A |  |
HUMAN:GATA3 | 11 | A | 0 | dvAGATAAvvd | HOCOMOCOv11 | ChIP-Seq | 0.901 | 0.893 | 98 | 60 | 500 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 4172 | 2625 | GATA3_HUMAN | P23771 |
| GATA4_HUMAN.H11MO.0.A |  |
HUMAN:GATA4 | 10 | A | 0 | dvWGATAASR | HOCOMOCOv11 | ChIP-Seq | 0.955 | 0.94 | 15 | 17 | 500 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 4173 | 2626 | GATA4_HUMAN | P43694 |
| GATA5_HUMAN.H11MO.0.D |  |
HUMAN:GATA5 | 17 | D | 0 | WvMnWGATAWbnhdRhd | HOCOMOCOv9 | Integrative | 13 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 15802 | 140628 | GATA5_HUMAN | Q9BWX5 | ||||
| GATA6_HUMAN.H11MO.0.A |  |
HUMAN:GATA6 | 13 | A | 0 | dvAGATAAGRddd | HOCOMOCOv11 | ChIP-Seq | 0.97 | 0.932 | 29 | 7 | 500 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 4174 | 2627 | GATA6_HUMAN | Q92908 |
| GBX1_HUMAN.H11MO.0.D |  |
HUMAN:GBX1 | 17 | D | 0 | bTAATTAvbnhhWWWdd | HOCOMOCOv10 | HT-SELEX | 38677 | HOX-related factors{3.1.1} | GBX (Gastrulation brain homeobox){3.1.1.11} | 4185 | 2636 | GBX1_HUMAN | Q14549 | ||||
| GBX2_HUMAN.H11MO.0.D | 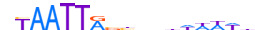 |
HUMAN:GBX2 | 17 | D | 0 | bYAATTRvbnhhhWWWd | HOCOMOCOv10 | HT-SELEX | 12613 | HOX-related factors{3.1.1} | GBX (Gastrulation brain homeobox){3.1.1.11} | 4186 | 2637 | GBX2_HUMAN | P52951 | ||||
| GCM1_HUMAN.H11MO.0.D |  |
HUMAN:GCM1 | 14 | D | 0 | hWnATGMKGGYhbb | HOCOMOCOv9 | Integrative | 12 | GCM factors{7.2.1} | GCMa (GCM1){7.2.1.0.1} | 4197 | 8521 | GCM1_HUMAN | Q9NP62 | ||||
| GCM2_HUMAN.H11MO.0.D |  |
HUMAN:GCM2 | 11 | D | 0 | bRTGCGGGTdn | HOCOMOCOv10 | HT-SELEX | 352 | GCM factors{7.2.1} | GCMb (GCM2){7.2.1.0.2} | 4198 | 9247 | GCM2_HUMAN | O75603 | ||||
| GCR_HUMAN.H11MO.0.A |  |
HUMAN:NR3C1 | 15 | A | 0 | dGdACWnAvWGTnCh | HOCOMOCOv11 | ChIP-Seq | 0.978 | 0.989 | 143 | 119 | 500 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 7978 | 2908 | GCR_HUMAN | P04150 |
| GCR_HUMAN.H11MO.1.A |  |
HUMAN:NR3C1 | 12 | A | 1 | hvRGRACAbdvh | HOCOMOCOv11 | ChIP-Seq | 0.93 | 0.925 | 143 | 119 | 500 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 7978 | 2908 | GCR_HUMAN | P04150 |
| GFI1B_HUMAN.H11MO.0.A |  |
HUMAN:GFI1B | 10 | A | 0 | RYWSWGATTK | HOCOMOCOv11 | ChIP-Seq | 0.891 | 0.887 | 3 | 24 | 503 | More than 3 adjacent zinc finger factors{2.3.3} | GFI1 factors{2.3.3.21} | 4238 | 8328 | GFI1B_HUMAN | Q5VTD9 |
| GFI1_HUMAN.H11MO.0.C |  |
HUMAN:GFI1 | 10 | C | 0 | RShSWGATTb | HOCOMOCOv9 | Integrative | 152 | More than 3 adjacent zinc finger factors{2.3.3} | GFI1 factors{2.3.3.21} | 4237 | 2672 | GFI1_HUMAN | Q99684 | ||||
| GLI1_HUMAN.H11MO.0.D |  |
HUMAN:GLI1 | 12 | D | 0 | bSTGGGTGGYCh | HOCOMOCOv11 | ChIP-Seq | 0.561 | 0.974 | 2 | 3 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 4317 | 2735 | GLI1_HUMAN | P08151 |
| GLI2_HUMAN.H11MO.0.D |  |
HUMAN:GLI2 | 11 | D | 0 | STGGGTGGTCb | HOCOMOCOv9 | Integrative | 970 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 4318 | 2736 | GLI2_HUMAN | P10070 | ||||
| GLI3_HUMAN.H11MO.0.B |  |
HUMAN:GLI3 | 11 | B | 0 | nTGGGTGGTYb | HOCOMOCOv9 | Integrative | 15 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 4319 | 2737 | GLI3_HUMAN | P10071 | ||||
| GLIS1_HUMAN.H11MO.0.D |  |
HUMAN:GLIS1 | 13 | D | 0 | hYGYGGGGGGTMd | HOCOMOCOv10 | HT-SELEX | 492 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 29525 | 148979 | GLIS1_HUMAN | Q8NBF1 | ||||
| GLIS2_HUMAN.H11MO.0.D |  |
HUMAN:GLIS2 | 14 | D | 0 | SdhYGYGGGGGGTv | HOCOMOCOv10 | HT-SELEX | 1681 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 29450 | 84662 | GLIS2_HUMAN | Q9BZE0 | ||||
| GLIS3_HUMAN.H11MO.0.D |  |
HUMAN:GLIS3 | 10 | D | 0 | SYGGGGGGTM | HOCOMOCOv9 | Integrative | 38 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 28510 | 169792 | GLIS3_HUMAN | Q8NEA6 | ||||
| GMEB2_HUMAN.H11MO.0.D |  |
HUMAN:GMEB2 | 11 | D | 0 | nndTACGTvvn | HOCOMOCOv10 | HT-SELEX | 4035 | GMEB{5.3.3} | GMEB{5.3.3.1} | 4371 | 26205 | GMEB2_HUMAN | Q9UKD1 | ||||
| GRHL1_HUMAN.H11MO.0.D |  |
HUMAN:GRHL1 | 12 | D | 0 | WWCCWGTYYbnY | HOCOMOCOv11 | ChIP-Seq | 0.731 | 3 | 500 | Grainyhead-related factors{6.7.1} | GRH-like proteins{6.7.1.1} | 17923 | 29841 | GRHL1_HUMAN | Q9NZI5 | ||
| GRHL2_HUMAN.H11MO.0.A |  |
HUMAN:GRHL2 | 12 | A | 0 | RWCYdGTYbddC | HOCOMOCOv11 | ChIP-Seq | 0.96 | 0.921 | 22 | 4 | 497 | Grainyhead-related factors{6.7.1} | GRH-like proteins{6.7.1.1} | 2799 | 79977 | GRHL2_HUMAN | Q6ISB3 |
| GSC2_HUMAN.H11MO.0.D |  |
HUMAN:GSC2 | 11 | D | 0 | ddvGGATKRRn | HOCOMOCOv10 | HT-SELEX | 610 | Paired-related HD factors{3.1.3} | GSC{3.1.3.9} | 4613 | 2928 | GSC2_HUMAN | O15499 | ||||
| GSC_HUMAN.H11MO.0.D |  |
HUMAN:GSC | 11 | D | 0 | ddvGGATTAdb | HOCOMOCOv10 | HT-SELEX | 1242 | Paired-related HD factors{3.1.3} | GSC{3.1.3.9} | 4612 | 145258 | GSC_HUMAN | P56915 | ||||
| GSX1_HUMAN.H11MO.0.D |  |
HUMAN:GSX1 | 15 | D | 0 | hRATKRvndddWdRK | HOCOMOCOv10 | HT-SELEX | 277 | HOX-related factors{3.1.1} | GSX{3.1.1.12} | 20374 | 219409 | GSX1_HUMAN | Q9H4S2 | ||||
| GSX2_HUMAN.H11MO.0.D |  |
HUMAN:GSX2 | 16 | D | 0 | bTAATKRvnddWWWAY | HOCOMOCOv10 | HT-SELEX | 6133 | HOX-related factors{3.1.1} | GSX{3.1.1.12} | 24959 | 170825 | GSX2_HUMAN | Q9BZM3 | ||||
| HAND1_HUMAN.H11MO.0.D |  |
HUMAN:HAND1 | 17 | D | 0 | ddbYCTGShhWnYvTCW | HOCOMOCOv11 | ChIP-Seq | 0.995 | 3 | 500 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 4807 | 9421 | HAND1_HUMAN | O96004 | ||
| HAND1_HUMAN.H11MO.1.D |  |
HUMAN:HAND1 | 7 | D | 1 | dSYCTGS | HOCOMOCOv11 | ChIP-Seq | 0.951 | 3 | 500 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 4807 | 9421 | HAND1_HUMAN | O96004 | ||
| HBP1_HUMAN.H11MO.0.D |  |
HUMAN:HBP1 | 9 | D | 0 | RnYSATTGA | HOCOMOCOv9 | Integrative | 5 | SOX-related factors{4.1.1} | Further Sox-related factors{4.1.1.9} | 23200 | 26959 | HBP1_HUMAN | O60381 | ||||
| HEN1_HUMAN.H11MO.0.C |  |
HUMAN:NHLH1 | 20 | C | 0 | bKGGnMbCAGCTGCGKCYhh | HOCOMOCOv9 | Integrative | 55 | Tal-related factors{1.2.3} | Tal / HEN-like factors{1.2.3.1} | 7817 | 4807 | HEN1_HUMAN | Q02575 | ||||
| HES1_HUMAN.H11MO.0.D |  |
HUMAN:HES1 | 13 | D | 0 | vMSvCAMKvGMhv | HOCOMOCOv9 | Integrative | 28 | Hairy-related factors{1.2.4} | Hairy-like factors{1.2.4.1} | 5192 | 3280 | HES1_HUMAN | Q14469 | ||||
| HES5_HUMAN.H11MO.0.D |  |
HUMAN:HES5 | 12 | D | 0 | hGGCACGTGYCd | HOCOMOCOv10 | HT-SELEX | 6731 | Hairy-related factors{1.2.4} | Hairy-like factors{1.2.4.1} | 19764 | 388585 | HES5_HUMAN | Q5TA89 | ||||
| HES7_HUMAN.H11MO.0.D |  |
HUMAN:HES7 | 12 | D | 0 | WGGCACGTGCCd | HOCOMOCOv10 | HT-SELEX | 379 | Hairy-related factors{1.2.4} | Hairy-like factors{1.2.4.1} | 15977 | 84667 | HES7_HUMAN | Q9BYE0 | ||||
| HESX1_HUMAN.H11MO.0.D |  |
HUMAN:HESX1 | 16 | D | 0 | nKCYGRCWCnTRRnhY | HOCOMOCOv9 | Integrative | 8 | Paired-related HD factors{3.1.3} | HESX{3.1.3.10} | 4877 | 8820 | HESX1_HUMAN | Q9UBX0 | ||||
| HEY1_HUMAN.H11MO.0.D |  |
HUMAN:HEY1 | 12 | D | 0 | bKRCACGTGbSd | HOCOMOCOv10 | HT-SELEX | 265 | Hairy-related factors{1.2.4} | Hairy-like factors{1.2.4.1} | 4880 | 23462 | HEY1_HUMAN | Q9Y5J3 | ||||
| HEY2_HUMAN.H11MO.0.D |  |
HUMAN:HEY2 | 17 | D | 0 | GKKSGCWCGTGSCnYKv | HOCOMOCOv9 | Integrative | 8 | Hairy-related factors{1.2.4} | Hairy-like factors{1.2.4.1} | 4881 | 23493 | HEY2_HUMAN | Q9UBP5 | ||||
| HIC1_HUMAN.H11MO.0.C |  |
HUMAN:HIC1 | 9 | C | 0 | dKGKTGCCM | HOCOMOCOv9 | Integrative | 40 | Factors with multiple dispersed zinc fingers{2.3.4} | Hypermethylated in Cancer proteins{2.3.4.17} | 4909 | 3090 | HIC1_HUMAN | Q14526 | ||||
| HIC2_HUMAN.H11MO.0.D |  |
HUMAN:HIC2 | 11 | D | 0 | nhRTGCCAGnn | HOCOMOCOv10 | HT-SELEX | 134 | Factors with multiple dispersed zinc fingers{2.3.4} | Hypermethylated in Cancer proteins{2.3.4.17} | 18595 | 23119 | HIC2_HUMAN | Q96JB3 | ||||
| HIF1A_HUMAN.H11MO.0.C |  |
HUMAN:HIF1A | 8 | C | 0 | vdACGTGh | HOCOMOCOv11 | ChIP-Seq | 0.762 | 0.738 | 39 | 8 | 501 | PAS domain factors{1.2.5} | Ahr-like factors{1.2.5.1} | 4910 | 3091 | HIF1A_HUMAN | Q16665 |
| HINFP_HUMAN.H11MO.0.C |  |
HUMAN:HINFP | 16 | C | 0 | dhSnnvdCGGACGTWv | HOCOMOCOv9 | Integrative | 42 | Factors with multiple dispersed zinc fingers{2.3.4} | HINFP-like factors{2.3.4.21} | 17850 | 25988 | HINFP_HUMAN | Q9BQA5 | ||||
| HLF_HUMAN.H11MO.0.C |  |
HUMAN:HLF | 13 | C | 0 | nRTKRYGTAAYvn | HOCOMOCOv9 | Integrative | 35 | C/EBP-related{1.1.8} | PAR factors{1.1.8.2} | 4977 | 3131 | HLF_HUMAN | Q16534 | ||||
| HLTF_HUMAN.H11MO.0.D |  |
HUMAN:HLTF | 12 | D | 0 | nAnGKCWGCAAM | HOCOMOCOv9 | Integrative | 10 | 11099 | 6596 | HLTF_HUMAN | Q14527 | ||||||
| HMBX1_HUMAN.H11MO.0.D |  |
HUMAN:HMBOX1 | 9 | D | 0 | dRYTRGTbA | HOCOMOCOv11 | ChIP-Seq | 0.76 | 7 | 500 | POU domain factors{3.1.10} | HNF1-like factors{3.1.10.7} | 26137 | 79618 | HMBX1_HUMAN | Q6NT76 | ||
| HME1_HUMAN.H11MO.0.D |  |
HUMAN:EN1 | 17 | D | 0 | vbTAATTAvbhhWWWWd | HOCOMOCOv10 | HT-SELEX | 817 | NK-related factors{3.1.2} | EN (Engrailed-like factors){3.1.2.7} | 3342 | 2019 | HME1_HUMAN | Q05925 | ||||
| HME2_HUMAN.H11MO.0.D |  |
HUMAN:EN2 | 11 | D | 0 | nnbRRTTRvnn | HOCOMOCOv10 | HT-SELEX | 9521 | NK-related factors{3.1.2} | EN (Engrailed-like factors){3.1.2.7} | 3343 | 2020 | HME2_HUMAN | P19622 | ||||
| HMGA1_HUMAN.H11MO.0.D |  |
HUMAN:HMGA1 | 7 | D | 0 | nWATTWd | HOCOMOCOv9 | Integrative | 97 | HMGA factors{8.2.1} | HMGA1 (HMGI(Y)){8.2.1.0.1} | 5010 | 3159 | HMGA1_HUMAN | P17096 | ||||
| HMGA2_HUMAN.H11MO.0.D |  |
HUMAN:HMGA2 | 15 | D | 0 | AATAWhSSSSAATAT | HOCOMOCOv9 | Integrative | 75 | HMGA factors{8.2.1} | HMGA2 (HMGI-C){8.2.1.0.2} | 5009 | 8091 | HMGA2_HUMAN | P52926 | ||||
| HMX1_HUMAN.H11MO.0.D |  |
HUMAN:HMX1 | 17 | D | 0 | dYTAATTGnYnnWTWAK | HOCOMOCOv10 | HT-SELEX | 34 | NK-related factors{3.1.2} | NK-5/HMX{3.1.2.18} | 5017 | 3166 | HMX1_HUMAN | Q9NP08 | ||||
| HMX2_HUMAN.H11MO.0.D |  |
HUMAN:HMX2 | 11 | D | 0 | nWTAAKTGnbh | HOCOMOCOv10 | HT-SELEX | 1221 | NK-related factors{3.1.2} | NK-5/HMX{3.1.2.18} | 5018 | 3167 | HMX2_HUMAN | A2RU54 | ||||
| HMX3_HUMAN.H11MO.0.D |  |
HUMAN:HMX3 | 11 | D | 0 | nnYTAATTGbh | HOCOMOCOv10 | HT-SELEX | 38692 | NK-related factors{3.1.2} | NK-5/HMX{3.1.2.18} | 5019 | 340784 | HMX3_HUMAN | A6NHT5 | ||||
| HNF1A_HUMAN.H11MO.0.C |  |
HUMAN:HNF1A | 15 | C | 0 | dRKWAATGWTTWRYh | HOCOMOCOv11 | ChIP-Seq | 0.927 | 3 | 500 | POU domain factors{3.1.10} | HNF1-like factors{3.1.10.7} | 11621 | 6927 | HNF1A_HUMAN | P20823 | ||
| HNF1B_HUMAN.H11MO.0.A |  |
HUMAN:HNF1B | 15 | A | 0 | dRTWAATGATTRMYh | HOCOMOCOv11 | ChIP-Seq | 0.95 | 0.994 | 5 | 8 | 500 | POU domain factors{3.1.10} | HNF1-like factors{3.1.10.7} | 11630 | 6928 | HNF1B_HUMAN | P35680 |
| HNF1B_HUMAN.H11MO.1.A |  |
HUMAN:HNF1B | 11 | A | 1 | dGTTAATnATT | HOCOMOCOv11 | ChIP-Seq | 0.916 | 0.993 | 5 | 8 | 500 | POU domain factors{3.1.10} | HNF1-like factors{3.1.10.7} | 11630 | 6928 | HNF1B_HUMAN | P35680 |
| HNF4A_HUMAN.H11MO.0.A |  |
HUMAN:HNF4A | 14 | A | 0 | vRGdnCAAAGTYCR | HOCOMOCOv11 | ChIP-Seq | 0.985 | 0.981 | 49 | 33 | 500 | RXR-related receptors (NR2){2.1.3} | HNF-4 (NR2A){2.1.3.2} | 5024 | 3172 | HNF4A_HUMAN | P41235 |
| HNF4G_HUMAN.H11MO.0.B |  |
HUMAN:HNF4G | 14 | B | 0 | vRGdnCAAAGKbCR | HOCOMOCOv11 | ChIP-Seq | 0.983 | 6 | 500 | RXR-related receptors (NR2){2.1.3} | HNF-4 (NR2A){2.1.3.2} | 5026 | 3174 | HNF4G_HUMAN | Q14541 | ||
| HNF6_HUMAN.H11MO.0.B |  |
HUMAN:ONECUT1 | 11 | B | 0 | hbYATTGATYd | HOCOMOCOv11 | ChIP-Seq | 0.976 | 24 | 512 | HD-CUT factors{3.1.9} | ONECUT{3.1.9.1} | 8138 | 3175 | HNF6_HUMAN | Q9UBC0 | ||
| HOMEZ_HUMAN.H11MO.0.D |  |
HUMAN:HOMEZ | 15 | D | 0 | ddWWATCGTTTWhhh | HOCOMOCOv10 | HT-SELEX | 275 | HD-ZF factors{3.1.8} | ZHX{3.1.8.5} | 20164 | 57594 | HOMEZ_HUMAN | Q8IX15 | ||||
| HSF1_HUMAN.H11MO.0.A |  |
HUMAN:HSF1 | 15 | A | 0 | bRGAAnbWWCYRGAR | HOCOMOCOv11 | ChIP-Seq | 0.997 | 0.984 | 56 | 32 | 510 | HSF factors{3.4.1} | HSF1 (HSTF1){3.4.1.0.1} | 5224 | 3297 | HSF1_HUMAN | Q00613 |
| HSF1_HUMAN.H11MO.1.A |  |
HUMAN:HSF1 | 9 | A | 1 | bKCTGGAAd | HOCOMOCOv11 | ChIP-Seq | 0.946 | 0.911 | 56 | 32 | 109 | HSF factors{3.4.1} | HSF1 (HSTF1){3.4.1.0.1} | 5224 | 3297 | HSF1_HUMAN | Q00613 |
| HSF2_HUMAN.H11MO.0.A |  |
HUMAN:HSF2 | 14 | A | 0 | vSRWbvWKSKvGRh | HOCOMOCOv9 | Integrative | 1043 | HSF factors{3.4.1} | HSF2 (HSTF2){3.4.1.0.2} | 5225 | 3298 | HSF2_HUMAN | Q03933 | ||||
| HSF4_HUMAN.H11MO.0.D |  |
HUMAN:HSF4 | 15 | D | 0 | hvGAAbvTTCYRGAW | HOCOMOCOv10 | HT-SELEX | 17390 | HSF factors{3.4.1} | HSF4 (HSTF4){3.4.1.0.3} | 5227 | 3299 | HSF4_HUMAN | Q9ULV5 | ||||
| HSFY1_HUMAN.H11MO.0.D |  |
HUMAN:HSFY1; HUMAN:HSFY2 | 12 | D | 0 | dWTMGAACGhdW | HOCOMOCOv10 | HT-SELEX | 6657 | HSF factors{3.4.1} | HSFY1 (HSFY2, HSF2L){3.4.1.0.6} | 18568; 23950 | 159119; 86614 | HSFY1_HUMAN | Q96LI6 | ||||
| HTF4_HUMAN.H11MO.0.A |  |
HUMAN:TCF12 | 10 | A | 0 | nSCAGSTGKS | HOCOMOCOv11 | ChIP-Seq | 0.915 | 0.987 | 49 | 10 | 502 | E2A-related factors{1.2.1} | HTF-4 (TCF-12, HEB){1.2.1.0.3} | 11623 | 6938 | HTF4_HUMAN | Q99081 |
| HXA10_HUMAN.H11MO.0.C |  |
HUMAN:HOXA10 | 12 | C | 0 | dnYnRTTTATGh | HOCOMOCOv9 | Integrative | 10 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5100 | 3206 | HXA10_HUMAN | P31260 | ||||
| HXA11_HUMAN.H11MO.0.D |  |
HUMAN:HOXA11 | 13 | D | 0 | WWWhnWTTTAhdd | HOCOMOCOv10 | HT-SELEX | 2642 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5101 | 3207 | HXA11_HUMAN | P31270 | ||||
| HXA13_HUMAN.H11MO.0.C |  |
HUMAN:HOXA13 | 11 | C | 0 | vdTWTTATKGS | HOCOMOCOv9 | Integrative | 18 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5102 | 3209 | HXA13_HUMAN | P31271 | ||||
| HXA1_HUMAN.H11MO.0.C |  |
HUMAN:HOXA1 | 10 | C | 0 | nTGATGGATG | HOCOMOCOv9 | Integrative | 8 | HOX-related factors{3.1.1} | HOX1{3.1.1.1} | 5099 | 3198 | HXA1_HUMAN | P49639 | ||||
| HXA2_HUMAN.H11MO.0.D |  |
HUMAN:HOXA2 | 11 | D | 0 | nvTMATTAdnd | HOCOMOCOv10 | HT-SELEX | 14435 | HOX-related factors{3.1.1} | HOX2{3.1.1.2} | 5103 | 3199 | HXA2_HUMAN | O43364 | ||||
| HXA5_HUMAN.H11MO.0.D |  |
HUMAN:HOXA5 | 10 | D | 0 | YTvATTARYR | HOCOMOCOv9 | Integrative | 54 | HOX-related factors{3.1.1} | HOX5{3.1.1.5} | 5106 | 3202 | HXA5_HUMAN | P20719 | ||||
| HXA7_HUMAN.H11MO.0.D | 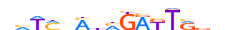 |
HUMAN:HOXA7 | 15 | D | 0 | vhTMnMnMGAKTRvv | HOCOMOCOv9 | Integrative | 21 | HOX-related factors{3.1.1} | HOX6-7{3.1.1.6} | 5108 | 3204 | HXA7_HUMAN | P31268 | ||||
| HXA9_HUMAN.H11MO.0.B |  |
HUMAN:HOXA9 | 11 | B | 0 | dTGATTKAYdd | HOCOMOCOv11 | ChIP-Seq | 0.883 | 0.757 | 5 | 8 | 500 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5109 | 3205 | HXA9_HUMAN | P31269 |
| HXB13_HUMAN.H11MO.0.A |  |
HUMAN:HOXB13 | 10 | A | 0 | YTTTATdRvn | HOCOMOCOv11 | ChIP-Seq | 0.846 | 9 | 500 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5112 | 10481 | HXB13_HUMAN | Q92826 | ||
| HXB1_HUMAN.H11MO.0.D |  |
HUMAN:HOXB1 | 10 | D | 0 | WGATGGATGG | HOCOMOCOv9 | Integrative | 15 | HOX-related factors{3.1.1} | HOX1{3.1.1.1} | 5111 | 3211 | HXB1_HUMAN | P14653 | ||||
| HXB2_HUMAN.H11MO.0.D |  |
HUMAN:HOXB2 | 15 | D | 0 | hvGAAbvTTCYAGAh | HOCOMOCOv10 | HT-SELEX | 420 | HOX-related factors{3.1.1} | HOX2{3.1.1.2} | 5113 | 3212 | HXB2_HUMAN | P14652 | ||||
| HXB3_HUMAN.H11MO.0.D |  |
HUMAN:HOXB3 | 14 | D | 0 | hTAATKRbhhhWWh | HOCOMOCOv10 | HT-SELEX | 3405 | HOX-related factors{3.1.1} | HOX3{3.1.1.3} | 5114 | 3213 | HXB3_HUMAN | P14651 | ||||
| HXB4_HUMAN.H11MO.0.B |  |
HUMAN:HOXB4 | 8 | B | 0 | bTAATKRS | HOCOMOCOv11 | ChIP-Seq | 0.491 | 0.894 | 2 | 12 | 502 | HOX-related factors{3.1.1} | HOX4{3.1.1.4} | 5115 | 3214 | HXB4_HUMAN | P17483 |
| HXB6_HUMAN.H11MO.0.D |  |
HUMAN:HOXB6 | 13 | D | 0 | dWTGATTKATGCn | HOCOMOCOv9 | Integrative | 7 | HOX-related factors{3.1.1} | HOX6-7{3.1.1.6} | 5117 | 3216 | HXB6_HUMAN | P17509 | ||||
| HXB7_HUMAN.H11MO.0.C |  |
HUMAN:HOXB7 | 10 | C | 0 | WTGATTRATK | HOCOMOCOv9 | Integrative | 28 | HOX-related factors{3.1.1} | HOX6-7{3.1.1.6} | 5118 | 3217 | HXB7_HUMAN | P09629 | ||||
| HXB8_HUMAN.H11MO.0.C |  |
HUMAN:HOXB8 | 11 | C | 0 | TTGATTRATKv | HOCOMOCOv9 | Integrative | 26 | HOX-related factors{3.1.1} | HOX8{3.1.1.7} | 5119 | 3218 | HXB8_HUMAN | P17481 | ||||
| HXC10_HUMAN.H11MO.0.D | 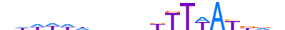 |
HUMAN:HOXC10 | 19 | D | 0 | ddWWWddnndKTTWAYddb | HOCOMOCOv10 | HT-SELEX | 9418 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5122 | 3226 | HXC10_HUMAN | Q9NYD6 | ||||
| HXC11_HUMAN.H11MO.0.D |  |
HUMAN:HOXC11 | 11 | D | 0 | dKTTWAYKRbh | HOCOMOCOv10 | HT-SELEX | 318 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5123 | 3227 | HXC11_HUMAN | O43248 | ||||
| HXC12_HUMAN.H11MO.0.D |  |
HUMAN:HOXC12 | 12 | D | 0 | dWTTTAYKRYbb | HOCOMOCOv10 | HT-SELEX | 2466 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5124 | 3228 | HXC12_HUMAN | P31275 | ||||
| HXC13_HUMAN.H11MO.0.D |  |
HUMAN:HOXC13 | 11 | D | 0 | ndTTTTACbvb | HOCOMOCOv10 | HT-SELEX | 1231 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5125 | 3229 | HXC13_HUMAN | P31276 | ||||
| HXC6_HUMAN.H11MO.0.D |  |
HUMAN:HOXC6 | 15 | D | 0 | WTGATTTATTWCTTT | HOCOMOCOv9 | Integrative | 7 | HOX-related factors{3.1.1} | HOX6-7{3.1.1.6} | 5128 | 3223 | HXC6_HUMAN | P09630 | ||||
| HXC8_HUMAN.H11MO.0.D |  |
HUMAN:HOXC8 | 14 | D | 0 | nTTSATTRAnKSnn | HOCOMOCOv9 | Integrative | 9 | HOX-related factors{3.1.1} | HOX8{3.1.1.7} | 5129 | 3224 | HXC8_HUMAN | P31273 | ||||
| HXC9_HUMAN.H11MO.0.C |  |
HUMAN:HOXC9 | 10 | C | 0 | hTTWATKGSh | HOCOMOCOv11 | ChIP-Seq | 0.871 | 2 | 500 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5130 | 3225 | HXC9_HUMAN | P31274 | ||
| HXD10_HUMAN.H11MO.0.D |  |
HUMAN:HOXD10 | 10 | D | 0 | TGnTYTWRTK | HOCOMOCOv9 | Integrative | 7 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5133 | 3236 | HXD10_HUMAN | P28358 | ||||
| HXD11_HUMAN.H11MO.0.D |  |
HUMAN:HOXD11 | 11 | D | 0 | dhTTTATKdnn | HOCOMOCOv10 | HT-SELEX | 124 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5134 | 3237 | HXD11_HUMAN | P31277 | ||||
| HXD12_HUMAN.H11MO.0.D |  |
HUMAN:HOXD12 | 11 | D | 0 | nnKTTTAnbdn | HOCOMOCOv10 | HT-SELEX | 8291 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5135 | 3238 | HXD12_HUMAN | P35452 | ||||
| HXD13_HUMAN.H11MO.0.D |  |
HUMAN:HOXD13 | 11 | D | 0 | WTTAYKRKRGM | HOCOMOCOv9 | Integrative | 13 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5136 | 3239 | HXD13_HUMAN | P35453 | ||||
| HXD3_HUMAN.H11MO.0.D |  |
HUMAN:HOXD3 | 11 | D | 0 | nRbTAATTRbd | HOCOMOCOv10 | HT-SELEX | 601 | HOX-related factors{3.1.1} | HOX3{3.1.1.3} | 5137 | 3232 | HXD3_HUMAN | P31249 | ||||
| HXD4_HUMAN.H11MO.0.D |  |
HUMAN:HOXD4 | 8 | D | 0 | KTAATTnW | HOCOMOCOv9 | Integrative | 8 | HOX-related factors{3.1.1} | HOX4{3.1.1.4} | 5138 | 3233 | HXD4_HUMAN | P09016 | ||||
| HXD8_HUMAN.H11MO.0.D |  |
HUMAN:HOXD8 | 11 | D | 0 | hhWTWTRCvnn | HOCOMOCOv10 | HT-SELEX | 278 | HOX-related factors{3.1.1} | HOX8{3.1.1.7} | 5139 | 3234 | HXD8_HUMAN | P13378 | ||||
| HXD9_HUMAN.H11MO.0.D |  |
HUMAN:HOXD9 | 10 | D | 0 | AnWTTnATnn | HOCOMOCOv9 | Integrative | 6 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 5140 | 3235 | HXD9_HUMAN | P28356 | ||||
| ID4_HUMAN.H11MO.0.D |  |
HUMAN:ID4 | 11 | D | 0 | nnCAGGTGhnn | HOCOMOCOv10 | HT-SELEX | 210 | HLH domain only{1.2.8} | Id4{1.2.8.0.4} | 5363 | 3400 | ID4_HUMAN | P47928 | ||||
| IKZF1_HUMAN.H11MO.0.C |  |
HUMAN:IKZF1 | 8 | C | 0 | bTGGGARd | HOCOMOCOv9 | Integrative | 61 | Factors with multiple dispersed zinc fingers{2.3.4} | Ikaros{2.3.4.4} | 13176 | 10320 | IKZF1_HUMAN | Q13422 | ||||
| INSM1_HUMAN.H11MO.0.C |  |
HUMAN:INSM1 | 12 | C | 0 | KGYMAGGGGGCA | HOCOMOCOv9 | Integrative | 24 | Factors with multiple dispersed zinc fingers{2.3.4} | Insulinoma-associated proteins{2.3.4.16} | 6090 | 3642 | INSM1_HUMAN | Q01101 | ||||
| IRF1_HUMAN.H11MO.0.A | 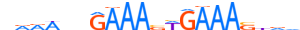 |
HUMAN:IRF1 | 20 | A | 0 | vRRWndGAAASWGAAAShvv | HOCOMOCOv11 | ChIP-Seq | 0.997 | 0.998 | 23 | 18 | 499 | Interferon-regulatory factors{3.5.3} | IRF-1{3.5.3.0.1} | 6116 | 3659 | IRF1_HUMAN | P10914 |
| IRF2_HUMAN.H11MO.0.A | 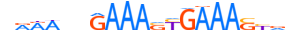 |
HUMAN:IRF2 | 20 | A | 0 | nRRWvhGAAASYGAAASYvv | HOCOMOCOv11 | ChIP-Seq | 0.994 | 8 | 499 | Interferon-regulatory factors{3.5.3} | IRF-2{3.5.3.0.2} | 6117 | 3660 | IRF2_HUMAN | P14316 | ||
| IRF3_HUMAN.H11MO.0.B |  |
HUMAN:IRF3 | 20 | B | 0 | dRRRddRAAAvdGRAASnRR | HOCOMOCOv11 | ChIP-Seq | 0.789 | 0.948 | 6 | 21 | 505 | Interferon-regulatory factors{3.5.3} | IRF-3{3.5.3.0.3} | 6118 | 3661 | IRF3_HUMAN | Q14653 |
| IRF4_HUMAN.H11MO.0.A |  |
HUMAN:IRF4 | 18 | A | 0 | vdWdRvGGAASTGARAvh | HOCOMOCOv11 | ChIP-Seq | 0.951 | 0.977 | 19 | 61 | 500 | Interferon-regulatory factors{3.5.3} | IRF-4 (LSIRF, NF-EM5, MUM1, Pip){3.5.3.0.4} | 6119 | 3662 | IRF4_HUMAN | Q15306 |
| IRF5_HUMAN.H11MO.0.D |  |
HUMAN:IRF5 | 20 | D | 0 | KAAAGGAAAGCnAAAAGTGM | HOCOMOCOv9 | Integrative | 5 | Interferon-regulatory factors{3.5.3} | IRF-5{3.5.3.0.5} | 6120 | 3663 | IRF5_HUMAN | Q13568 | ||||
| IRF7_HUMAN.H11MO.0.C | 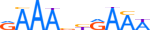 |
HUMAN:IRF7 | 10 | C | 0 | RAAAbYRAAW | HOCOMOCOv9 | Integrative | 40 | Interferon-regulatory factors{3.5.3} | IRF-7{3.5.3.0.7} | 6122 | 3665 | IRF7_HUMAN | Q92985 | ||||
| IRF8_HUMAN.H11MO.0.B | 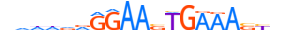 |
HUMAN:IRF8 | 20 | B | 0 | vdddvRGGAASTGAAASYdv | HOCOMOCOv11 | ChIP-Seq | 0.996 | 59 | 505 | Interferon-regulatory factors{3.5.3} | IRF-8 (ICSBP1){3.5.3.0.8} | 5358 | 3394 | IRF8_HUMAN | Q02556 | ||
| IRF9_HUMAN.H11MO.0.C |  |
HUMAN:IRF9 | 12 | C | 0 | GAAAbhGAAACT | HOCOMOCOv9 | Integrative | 5 | Interferon-regulatory factors{3.5.3} | IRF-9 (ISGF-3gamma){3.5.3.0.9} | 6131 | 10379 | IRF9_HUMAN | Q00978 | ||||
| IRX2_HUMAN.H11MO.0.D |  |
HUMAN:IRX2 | 19 | D | 0 | vbdbKdndKKdYRTGKdvd | HOCOMOCOv10 | HT-SELEX | 19310 | TALE-type homeo domain factors{3.1.4} | IRX (Iroquois){3.1.4.1} | 14359 | 153572 | IRX2_HUMAN | Q9BZI1 | ||||
| IRX3_HUMAN.H11MO.0.D |  |
HUMAN:IRX3 | 11 | D | 0 | hndYWTGTWhd | HOCOMOCOv10 | HT-SELEX | 383 | TALE-type homeo domain factors{3.1.4} | IRX (Iroquois){3.1.4.1} | 14360 | 79191 | IRX3_HUMAN | P78415 | ||||
| ISL1_HUMAN.H11MO.0.A |  |
HUMAN:ISL1 | 9 | A | 0 | vYWRAKKRv | HOCOMOCOv11 | ChIP-Seq | 0.805 | 0.858 | 3 | 13 | 500 | HD-LIM factors{3.1.5} | ISL{3.1.5.1} | 6132 | 3670 | ISL1_HUMAN | P61371 |
| ISL2_HUMAN.H11MO.0.D |  |
HUMAN:ISL2 | 11 | D | 0 | hnbTAAKTGvh | HOCOMOCOv10 | HT-SELEX | 16988 | HD-LIM factors{3.1.5} | ISL{3.1.5.1} | 18524 | 64843 | ISL2_HUMAN | Q96A47 | ||||
| ISX_HUMAN.H11MO.0.D |  |
HUMAN:ISX | 16 | D | 0 | bTAATTARnhhWhhdd | HOCOMOCOv10 | HT-SELEX | 14859 | Paired-related HD factors{3.1.3} | ISX{3.1.3.12} | 28084 | 91464 | ISX_HUMAN | Q2M1V0 | ||||
| ITF2_HUMAN.H11MO.0.C |  |
HUMAN:TCF4 | 10 | C | 0 | dSCAGSTGbb | HOCOMOCOv11 | ChIP-Seq | 0.949 | 4 | 499 | E2A-related factors{1.2.1} | SEF2 (E2-2, TCF-4, ITF-2){1.2.1.0.2} | 11634 | 6925 | ITF2_HUMAN | P15884 | ||
| JDP2_HUMAN.H11MO.0.D |  |
HUMAN:JDP2 | 12 | D | 0 | bRATGACGTCAb | HOCOMOCOv10 | HT-SELEX | 2851 | Fos-related factors{1.1.2} | ATF-3-like factors{1.1.2.2} | 17546 | 122953 | JDP2_HUMAN | Q8WYK2 | ||||
| JUNB_HUMAN.H11MO.0.A |  |
HUMAN:JUNB | 11 | A | 0 | dvTGAGTCAbh | HOCOMOCOv11 | ChIP-Seq | 0.977 | 0.919 | 8 | 25 | 500 | Jun-related factors{1.1.1} | Jun factors{1.1.1.1} | 6205 | 3726 | JUNB_HUMAN | P17275 |
| JUND_HUMAN.H11MO.0.A |  |
HUMAN:JUND | 11 | A | 0 | dvTGAGTCAbb | HOCOMOCOv11 | ChIP-Seq | 0.996 | 0.987 | 64 | 21 | 450 | Jun-related factors{1.1.1} | Jun factors{1.1.1.1} | 6206 | 3727 | JUND_HUMAN | P17535 |
| JUN_HUMAN.H11MO.0.A |  |
HUMAN:JUN | 11 | A | 0 | dvTGAGTCAYh | HOCOMOCOv11 | ChIP-Seq | 0.954 | 0.935 | 56 | 39 | 500 | Jun-related factors{1.1.1} | Jun factors{1.1.1.1} | 6204 | 3725 | JUN_HUMAN | P05412 |
| KAISO_HUMAN.H11MO.0.A |  |
HUMAN:ZBTB33 | 15 | A | 0 | vdRvbMYCGCGAGAv | HOCOMOCOv11 | ChIP-Seq | 0.9 | 37 | 498 | Other factors with up to three adjacent zinc fingers{2.3.2} | Factors with 2-3 adjacent zinc fingersand a BTB/POZ domain{2.3.2.1} | 16682 | 10009 | KAISO_HUMAN | Q86T24 | ||
| KAISO_HUMAN.H11MO.1.A |  |
HUMAN:ZBTB33 | 11 | A | 1 | hvYCGCGAGAn | HOCOMOCOv11 | ChIP-Seq | 0.888 | 37 | 502 | Other factors with up to three adjacent zinc fingers{2.3.2} | Factors with 2-3 adjacent zinc fingersand a BTB/POZ domain{2.3.2.1} | 16682 | 10009 | KAISO_HUMAN | Q86T24 | ||
| KAISO_HUMAN.H11MO.2.A |  |
HUMAN:ZBTB33 | 12 | A | 2 | hTvGCARGAddn | HOCOMOCOv9 | Integrative | 0.841 | 37 | 2038 | Other factors with up to three adjacent zinc fingers{2.3.2} | Factors with 2-3 adjacent zinc fingersand a BTB/POZ domain{2.3.2.1} | 16682 | 10009 | KAISO_HUMAN | Q86T24 | ||
| KLF12_HUMAN.H11MO.0.C |  |
HUMAN:KLF12 | 11 | C | 0 | vRGGGCGGGGC | HOCOMOCOv11 | ChIP-Seq | 0.808 | 3 | 452 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 6346 | 11278 | KLF12_HUMAN | Q9Y4X4 | ||
| KLF13_HUMAN.H11MO.0.D |  |
HUMAN:KLF13 | 11 | D | 0 | dRGKGGGCGdn | HOCOMOCOv10 | HT-SELEX | 7671 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 13672 | 51621 | KLF13_HUMAN | Q9Y2Y9 | ||||
| KLF14_HUMAN.H11MO.0.D |  |
HUMAN:KLF14 | 13 | D | 0 | ddGKGGGCGTGdb | HOCOMOCOv10 | HT-SELEX | 2224 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 23025 | KLF14_HUMAN | Q8TD94 | |||||
| KLF15_HUMAN.H11MO.0.A |  |
HUMAN:KLF15 | 19 | A | 0 | RRGGMGGRGvnRGGRRdSR | HOCOMOCOv11 | ChIP-Seq | 0.952 | 0.998 | 4 | 3 | 481 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 14536 | 28999 | KLF15_HUMAN | Q9UIH9 |
| KLF16_HUMAN.H11MO.0.D | 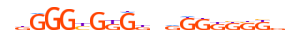 |
HUMAN:KLF16 | 19 | D | 0 | dbGGGYGKGdndGKKKSKb | HOCOMOCOv10 | HT-SELEX | 650 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 16857 | 83855 | KLF16_HUMAN | Q9BXK1 | ||||
| KLF1_HUMAN.H11MO.0.A | 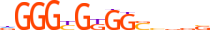 |
HUMAN:KLF1 | 14 | A | 0 | dGGGYGKGGShbbS | HOCOMOCOv11 | ChIP-Seq | 0.944 | 0.973 | 5 | 6 | 492 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 6345 | 10661 | KLF1_HUMAN | Q13351 |
| KLF3_HUMAN.H11MO.0.B |  |
HUMAN:KLF3 | 19 | B | 0 | vvvvvdGGGYGGRGbhbbv | HOCOMOCOv11 | ChIP-Seq | 0.76 | 0.992 | 2 | 7 | 498 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 16516 | 51274 | KLF3_HUMAN | P57682 |
| KLF4_HUMAN.H11MO.0.A |  |
HUMAN:KLF4 | 10 | A | 0 | dGGGYGKGGY | HOCOMOCOv11 | ChIP-Seq | 0.888 | 0.983 | 11 | 34 | 500 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 6348 | 9314 | KLF4_HUMAN | O43474 |
| KLF5_HUMAN.H11MO.0.A |  |
HUMAN:KLF5 | 14 | A | 0 | dGGGYGKGGYnKdn | HOCOMOCOv11 | ChIP-Seq | 0.969 | 0.974 | 32 | 23 | 502 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 6349 | 688 | KLF5_HUMAN | Q13887 |
| KLF6_HUMAN.H11MO.0.A |  |
HUMAN:KLF6 | 19 | A | 0 | vvvRGGGYGKRGvnbRSvv | HOCOMOCOv11 | ChIP-Seq | 0.946 | 0.604 | 16 | 2 | 498 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 2235 | 1316 | KLF6_HUMAN | Q99612 |
| KLF8_HUMAN.H11MO.0.C |  |
HUMAN:KLF8 | 9 | C | 0 | CAGGGGGTG | HOCOMOCOv9 | Integrative | 5 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 6351 | 11279 | KLF8_HUMAN | O95600 | ||||
| KLF9_HUMAN.H11MO.0.C |  |
HUMAN:KLF9 | 15 | C | 0 | SvvGKGGGCGTGKCb | HOCOMOCOv11 | ChIP-Seq | 0.954 | 3 | 256 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1123 | 687 | KLF9_HUMAN | Q13886 | ||
| LBX2_HUMAN.H11MO.0.D |  |
HUMAN:LBX2 | 11 | D | 0 | vYTAATTRGnb | HOCOMOCOv10 | HT-SELEX | 1425 | NK-related factors{3.1.2} | LBX{3.1.2.10} | 15525 | 85474 | LBX2_HUMAN | Q6XYB7 | ||||
| LEF1_HUMAN.H11MO.0.A |  |
HUMAN:LEF1 | 14 | A | 0 | bSYTTTSWhnKbhh | HOCOMOCOv11 | ChIP-Seq | 0.876 | 20 | 456 | TCF-7-related factors{4.1.3} | LEF-1 (TCF-1alpha) [1]{4.1.3.0.4} | 6551 | 51176 | LEF1_HUMAN | Q9UJU2 | ||
| LHX2_HUMAN.H11MO.0.A |  |
HUMAN:LHX2 | 12 | A | 0 | bbYTAATTRvhb | HOCOMOCOv11 | ChIP-Seq | 0.826 | 0.931 | 5 | 12 | 430 | HD-LIM factors{3.1.5} | Lhx-2-like factors{3.1.5.3} | 6594 | 9355 | LHX2_HUMAN | P50458 |
| LHX3_HUMAN.H11MO.0.C |  |
HUMAN:LHX3 | 13 | C | 0 | ddAWTTAATTAdh | HOCOMOCOv9 | Integrative | 72 | HD-LIM factors{3.1.5} | Lhx-3-like factors{3.1.5.4} | 6595 | 8022 | LHX3_HUMAN | Q9UBR4 | ||||
| LHX4_HUMAN.H11MO.0.D |  |
HUMAN:LHX4 | 11 | D | 0 | dnvYTAATKdn | HOCOMOCOv10 | HT-SELEX | 1423 | HD-LIM factors{3.1.5} | Lhx-3-like factors{3.1.5.4} | 21734 | 89884 | LHX4_HUMAN | Q969G2 | ||||
| LHX6_HUMAN.H11MO.0.D |  |
HUMAN:LHX6 | 10 | D | 0 | nnYTGATTRv | HOCOMOCOv11 | ChIP-Seq | 0.567 | 0.95 | 1 | 2 | 444 | HD-LIM factors{3.1.5} | Lhx-6-like factors{3.1.5.5} | 21735 | 26468 | LHX6_HUMAN | Q9UPM6 |
| LHX8_HUMAN.H11MO.0.D |  |
HUMAN:LHX8 | 18 | D | 0 | hvYTAATTRShvhYWWdd | HOCOMOCOv10 | HT-SELEX | 9063 | HD-LIM factors{3.1.5} | Lhx-6-like factors{3.1.5.5} | 28838 | 431707 | LHX8_HUMAN | Q68G74 | ||||
| LHX9_HUMAN.H11MO.0.D | 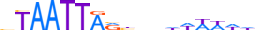 |
HUMAN:LHX9 | 18 | D | 0 | bTAATTARbnhhWWWWdd | HOCOMOCOv10 | HT-SELEX | 4409 | HD-LIM factors{3.1.5} | Lhx-2-like factors{3.1.5.3} | 14222 | 56956 | LHX9_HUMAN | Q9NQ69 | ||||
| LMX1A_HUMAN.H11MO.0.D |  |
HUMAN:LMX1A | 19 | D | 0 | YAATTAAWWnKnYWATTWn | HOCOMOCOv10 | HT-SELEX | 23 | HD-LIM factors{3.1.5} | Lmx{3.1.5.6} | 6653 | 4009 | LMX1A_HUMAN | Q8TE12 | ||||
| LMX1B_HUMAN.H11MO.0.D |  |
HUMAN:LMX1B | 18 | D | 0 | TAATTAAYWvvbWhWWWW | HOCOMOCOv10 | HT-SELEX | 35 | HD-LIM factors{3.1.5} | Lmx{3.1.5.6} | 6654 | 4010 | LMX1B_HUMAN | O60663 | ||||
| LYL1_HUMAN.H11MO.0.A |  |
HUMAN:LYL1 | 14 | A | 0 | vMWbMTGYbhYbbb | HOCOMOCOv11 | ChIP-Seq | 0.852 | 0.808 | 23 | 3 | 453 | Tal-related factors{1.2.3} | Tal / HEN-like factors{1.2.3.1} | 6734 | 4066 | LYL1_HUMAN | P12980 |
| MAFA_HUMAN.H11MO.0.D |  |
HUMAN:MAFA | 21 | D | 0 | bKGCTGWMShhSYWvYYWYYv | HOCOMOCOv9 | Integrative | 27 | Maf-related factors{1.1.3} | Large Maf factors{1.1.3.1} | 23145 | 389692 | MAFA_HUMAN | Q8NHW3 | ||||
| MAFB_HUMAN.H11MO.0.B |  |
HUMAN:MAFB | 11 | B | 0 | TGCTGAbhhdd | HOCOMOCOv11 | ChIP-Seq | 0.705 | 0.953 | 5 | 3 | 474 | Maf-related factors{1.1.3} | Large Maf factors{1.1.3.1} | 6408 | 9935 | MAFB_HUMAN | Q9Y5Q3 |
| MAFF_HUMAN.H11MO.0.B | 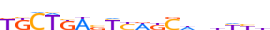 |
HUMAN:MAFF | 18 | B | 0 | KKCTKWbYYWKSdnhYYb | HOCOMOCOv11 | ChIP-Seq | 0.544 | 0.945 | 2 | 15 | 473 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 6780 | 23764 | MAFF_HUMAN | Q9ULX9 |
| MAFF_HUMAN.H11MO.1.B |  |
HUMAN:MAFF | 9 | B | 1 | TGCTGASYY | HOCOMOCOv11 | ChIP-Seq | 0.56 | 0.94 | 2 | 15 | 497 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 6780 | 23764 | MAFF_HUMAN | Q9ULX9 |
| MAFG_HUMAN.H11MO.0.A |  |
HUMAN:MAFG | 18 | A | 0 | ddWWnTGCTGASTCAGCW | HOCOMOCOv11 | ChIP-Seq | 0.869 | 0.998 | 3 | 6 | 430 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 6781 | 4097 | MAFG_HUMAN | O15525 |
| MAFG_HUMAN.H11MO.1.A |  |
HUMAN:MAFG | 8 | A | 1 | TGCTGWSK | HOCOMOCOv11 | ChIP-Seq | 0.874 | 0.989 | 3 | 6 | 500 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 6781 | 4097 | MAFG_HUMAN | O15525 |
| MAFK_HUMAN.H11MO.0.A |  |
HUMAN:MAFK | 18 | A | 0 | ddWWnTGCTGASTCAKSW | HOCOMOCOv11 | ChIP-Seq | 0.986 | 0.983 | 12 | 11 | 431 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 6782 | 7975 | MAFK_HUMAN | O60675 |
| MAFK_HUMAN.H11MO.1.A |  |
HUMAN:MAFK | 13 | A | 1 | WGCTGAGTMAKSW | HOCOMOCOv11 | ChIP-Seq | 0.961 | 0.98 | 12 | 11 | 410 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 6782 | 7975 | MAFK_HUMAN | O60675 |
| MAF_HUMAN.H11MO.0.A |  |
HUMAN:MAF | 19 | A | 0 | vdWdnTGCTGAbdhddvhd | HOCOMOCOv11 | ChIP-Seq | 0.853 | 0.837 | 10 | 5 | 500 | Maf-related factors{1.1.3} | Large Maf factors{1.1.3.1} | 6776 | 4094 | MAF_HUMAN | O75444 |
| MAF_HUMAN.H11MO.1.B |  |
HUMAN:MAF | 8 | B | 1 | WGCTGAbh | HOCOMOCOv11 | ChIP-Seq | 0.788 | 0.849 | 10 | 5 | 496 | Maf-related factors{1.1.3} | Large Maf factors{1.1.3.1} | 6776 | 4094 | MAF_HUMAN | O75444 |
| MAX_HUMAN.H11MO.0.A |  |
HUMAN:MAX | 10 | A | 0 | vnSCACGYGK | HOCOMOCOv11 | ChIP-Seq | 0.95 | 0.947 | 86 | 14 | 501 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 6913 | 4149 | MAX_HUMAN | P61244 |
| MAZ_HUMAN.H11MO.0.A |  |
HUMAN:MAZ | 22 | A | 0 | vRRGGWGGRRSnRRRRRvdvdR | HOCOMOCOv11 | ChIP-Seq | 0.951 | 0.961 | 3 | 5 | 501 | Factors with multiple dispersed zinc fingers{2.3.4} | MAZ-like factors{2.3.4.8} | 6914 | 4150 | MAZ_HUMAN | P56270 |
| MAZ_HUMAN.H11MO.1.A |  |
HUMAN:MAZ | 11 | A | 1 | RRGGGMGGGGv | HOCOMOCOv11 | ChIP-Seq | 0.926 | 0.955 | 3 | 5 | 486 | Factors with multiple dispersed zinc fingers{2.3.4} | MAZ-like factors{2.3.4.8} | 6914 | 4150 | MAZ_HUMAN | P56270 |
| MBD2_HUMAN.H11MO.0.B |  |
HUMAN:MBD2 | 11 | B | 0 | nSGGCCGGMKv | HOCOMOCOv9 | Integrative | 113 | 6917 | 8932 | MBD2_HUMAN | Q9UBB5 | ||||||
| MCR_HUMAN.H11MO.0.D |  |
HUMAN:NR3C2 | 17 | D | 0 | hAGWMCARSTKGTdGKR | HOCOMOCOv9 | Integrative | 8 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 7979 | 4306 | MCR_HUMAN | P08235 | ||||
| MECP2_HUMAN.H11MO.0.C |  |
HUMAN:MECP2 | 7 | C | 0 | SCCGGAG | HOCOMOCOv9 | Integrative | 122 | 6990 | 4204 | MECP2_HUMAN | P51608 | ||||||
| MEF2A_HUMAN.H11MO.0.A |  |
HUMAN:MEF2A | 13 | A | 0 | ddYTATTTWWRRh | HOCOMOCOv11 | ChIP-Seq | 0.946 | 0.973 | 15 | 6 | 500 | Regulators of differentiation{5.1.1} | MEF-2{5.1.1.1} | 6993 | 4205 | MEF2A_HUMAN | Q02078 |
| MEF2B_HUMAN.H11MO.0.A |  |
HUMAN:MEF2B | 14 | A | 0 | ddddYTATTTWWRK | HOCOMOCOv11 | ChIP-Seq | 0.946 | 23 | 500 | Regulators of differentiation{5.1.1} | MEF-2{5.1.1.1} | 6995 | 100271849; 4207 | MEF2B_HUMAN | Q02080 | ||
| MEF2C_HUMAN.H11MO.0.A |  |
HUMAN:MEF2C | 13 | A | 0 | ddYTATTTWWRRh | HOCOMOCOv11 | ChIP-Seq | 0.959 | 0.877 | 6 | 9 | 500 | Regulators of differentiation{5.1.1} | MEF-2{5.1.1.1} | 6996 | 4208 | MEF2C_HUMAN | Q06413 |
| MEF2D_HUMAN.H11MO.0.A |  |
HUMAN:MEF2D | 12 | A | 0 | dKYTATTTWTAR | HOCOMOCOv11 | ChIP-Seq | 0.843 | 0.965 | 2 | 27 | 500 | Regulators of differentiation{5.1.1} | MEF-2{5.1.1.1} | 6997 | 4209 | MEF2D_HUMAN | Q14814 |
| MEIS1_HUMAN.H11MO.0.A |  |
HUMAN:MEIS1 | 12 | A | 0 | dTGATWdATddb | HOCOMOCOv11 | ChIP-Seq | 0.816 | 0.945 | 2 | 37 | 499 | TALE-type homeo domain factors{3.1.4} | MEIS{3.1.4.2} | 7000 | 4211 | MEIS1_HUMAN | O00470 |
| MEIS1_HUMAN.H11MO.1.B |  |
HUMAN:MEIS1 | 9 | B | 1 | dhTGACARv | HOCOMOCOv11 | ChIP-Seq | 0.778 | 0.854 | 2 | 37 | 500 | TALE-type homeo domain factors{3.1.4} | MEIS{3.1.4.2} | 7000 | 4211 | MEIS1_HUMAN | O00470 |
| MEIS2_HUMAN.H11MO.0.B |  |
HUMAN:MEIS2 | 13 | B | 0 | hTGACAGbTKTbv | HOCOMOCOv9 | Integrative | 413 | TALE-type homeo domain factors{3.1.4} | MEIS{3.1.4.2} | 7001 | 4212 | MEIS2_HUMAN | O14770 | ||||
| MEIS3_HUMAN.H11MO.0.D |  |
HUMAN:MEIS3 | 11 | D | 0 | ndTGACAKvYd | HOCOMOCOv10 | HT-SELEX | 10453 | TALE-type homeo domain factors{3.1.4} | MEIS{3.1.4.2} | 29537 | 56917 | MEIS3_HUMAN | Q99687 | ||||
| MEOX1_HUMAN.H11MO.0.D | 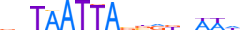 |
HUMAN:MEOX1 | 16 | D | 0 | nbTAATTAnbRbhWWh | HOCOMOCOv10 | HT-SELEX | 61 | HOX-related factors{3.1.1} | MEOX{3.1.1.14} | 7013 | 4222 | MEOX1_HUMAN | P50221 | ||||
| MEOX2_HUMAN.H11MO.0.D |  |
HUMAN:MEOX2 | 11 | D | 0 | hTAATTAbndh | HOCOMOCOv10 | HT-SELEX | 40067 | HOX-related factors{3.1.1} | MEOX{3.1.1.14} | 7014 | 4223 | MEOX2_HUMAN | P50222 | ||||
| MESP1_HUMAN.H11MO.0.D |  |
HUMAN:MESP1 | 11 | D | 0 | nCAGGTGbdnn | HOCOMOCOv10 | HT-SELEX | 61 | Tal-related factors{1.2.3} | Mesp-like factors{1.2.3.3} | 29658 | 55897 | MESP1_HUMAN | Q9BRJ9 | ||||
| MGAP_HUMAN.H11MO.0.D |  |
HUMAN:MGA | 16 | D | 0 | RRGTGTKAnWTWTMRC | HOCOMOCOv10 | HT-SELEX | 6178 | bHLH-ZIP factors{1.2.6}; TBX6-related factors{6.5.5} |
Mad-like factors{1.2.6.7}; MGA (MAD5){6.5.5.0.2} |
14010 | 23269 | MGAP_HUMAN | Q8IWI9 | ||||
| MITF_HUMAN.H11MO.0.A |  |
HUMAN:MITF | 10 | A | 0 | YCACGTGACh | HOCOMOCOv11 | ChIP-Seq | 0.999 | 0.824 | 28 | 3 | 516 | bHLH-ZIP factors{1.2.6} | TFE3-like factors{1.2.6.1} | 7105 | 4286 | MITF_HUMAN | O75030 |
| MIXL1_HUMAN.H11MO.0.D |  |
HUMAN:MIXL1 | 11 | D | 0 | vbTAATTvvdn | HOCOMOCOv10 | HT-SELEX | 220 | Paired-related HD factors{3.1.3} | MIX{3.1.3.14} | 13363 | 83881 | MIXL1_HUMAN | Q9H2W2 | ||||
| MLXPL_HUMAN.H11MO.0.D |  |
HUMAN:MLXIPL | 19 | D | 0 | SCAYGKGRMMMYnbSGTGb | HOCOMOCOv9 | Integrative | 11 | bHLH-ZIP factors{1.2.6} | Mondo-like factors{1.2.6.6} | 12744 | 51085 | MLXPL_HUMAN | Q9NP71 | ||||
| MLX_HUMAN.H11MO.0.D | 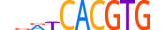 |
HUMAN:MLX | 11 | D | 0 | hdRWCACGTGn | HOCOMOCOv10 | HT-SELEX | 18958 | bHLH-ZIP factors{1.2.6} | Mondo-like factors{1.2.6.6} | 11645 | 6945 | MLX_HUMAN | Q9UH92 | ||||
| MNX1_HUMAN.H11MO.0.D |  |
HUMAN:MNX1 | 19 | D | 0 | hbWYTYdMnWhWCYhYnhh | HOCOMOCOv10 | HT-SELEX | 27 | HOX-related factors{3.1.1} | MNX{3.1.1.13} | 4979 | 3110 | MNX1_HUMAN | P50219 | ||||
| MSX1_HUMAN.H11MO.0.D |  |
HUMAN:MSX1 | 18 | D | 0 | WWATTRbdhbhTAATTRb | HOCOMOCOv10 | HT-SELEX | 34415 | NK-related factors{3.1.2} | MSX{3.1.2.11} | 7391 | 4487 | MSX1_HUMAN | P28360 | ||||
| MSX2_HUMAN.H11MO.0.D |  |
HUMAN:MSX2 | 7 | D | 0 | TAATTnb | HOCOMOCOv9 | Integrative | 12 | NK-related factors{3.1.2} | MSX{3.1.2.11} | 7392 | 4488 | MSX2_HUMAN | P35548 | ||||
| MTF1_HUMAN.H11MO.0.C |  |
HUMAN:MTF1 | 17 | C | 0 | vGKRMCGKGTGCAdMdb | HOCOMOCOv9 | Integrative | 33 | More than 3 adjacent zinc finger factors{2.3.3} | MTF1-like factors{2.3.3.24} | 7428 | 4520 | MTF1_HUMAN | Q14872 | ||||
| MXI1_HUMAN.H11MO.0.A | 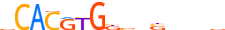 |
HUMAN:MXI1 | 15 | A | 0 | vCACRYGKvnSbbbv | HOCOMOCOv10 | ChIP-Seq | 0.878 | 0.973 | 10 | 5 | 500 | bHLH-ZIP factors{1.2.6} | Mad-like factors{1.2.6.7} | 7534 | 4601 | MXI1_HUMAN | P50539 |
| MXI1_HUMAN.H11MO.1.A |  |
HUMAN:MXI1 | 11 | A | 1 | SMCACRTGbhb | HOCOMOCOv11 | ChIP-Seq | 0.871 | 0.96 | 10 | 5 | 500 | bHLH-ZIP factors{1.2.6} | Mad-like factors{1.2.6.7} | 7534 | 4601 | MXI1_HUMAN | P50539 |
| MYBA_HUMAN.H11MO.0.D |  |
HUMAN:MYBL1 | 15 | D | 0 | vbbGRCvGTTRvnvv | HOCOMOCOv11 | ChIP-Seq | 0.533 | 0.897 | 3 | 2 | 500 | Myb/SANT domain factors{3.5.1} | Myb-like factors{3.5.1.1} | 7547 | 4603 | MYBA_HUMAN | P10243 |
| MYBB_HUMAN.H11MO.0.D |  |
HUMAN:MYBL2 | 10 | D | 0 | dRSAnGTTdW | HOCOMOCOv9 | Integrative | 9 | Myb/SANT domain factors{3.5.1} | Myb-like factors{3.5.1.1} | 7548 | 4605 | MYBB_HUMAN | P10244 | ||||
| MYB_HUMAN.H11MO.0.A |  |
HUMAN:MYB | 12 | A | 0 | dvhRRCMGYTRv | HOCOMOCOv11 | ChIP-Seq | 0.829 | 0.919 | 27 | 9 | 500 | Myb/SANT domain factors{3.5.1} | Myb-like factors{3.5.1.1} | 7545 | 4602 | MYB_HUMAN | P10242 |
| MYCN_HUMAN.H11MO.0.A |  |
HUMAN:MYCN | 12 | A | 0 | vRvCACGTGbdv | HOCOMOCOv11 | ChIP-Seq | 0.919 | 0.899 | 32 | 10 | 501 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 7559 | 4613 | MYCN_HUMAN | P04198 |
| MYC_HUMAN.H11MO.0.A |  |
HUMAN:MYC | 11 | A | 0 | vMCACGTGShb | HOCOMOCOv11 | ChIP-Seq | 0.947 | 0.948 | 265 | 126 | 501 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 7553 | 4609 | MYC_HUMAN | P01106 |
| MYF6_HUMAN.H11MO.0.C |  |
HUMAN:MYF6 | 7 | C | 0 | CAGCTGC | HOCOMOCOv9 | Integrative | 9 | MyoD / ASC-related factors{1.2.2} | Myogenic transcription factors{1.2.2.1} | 7566 | 4618 | MYF6_HUMAN | P23409 | ||||
| MYNN_HUMAN.H11MO.0.D |  |
HUMAN:MYNN | 17 | D | 0 | dRRCTYTTAWKbYRdhR | HOCOMOCOv11 | ChIP-Seq | 0.796 | 4 | 366 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 14955 | 55892 | MYNN_HUMAN | Q9NPC7 | ||
| MYOD1_HUMAN.H11MO.0.A |  |
HUMAN:MYOD1 | 15 | A | 0 | vCAGCTGYYnYbbbh | HOCOMOCOv11 | ChIP-Seq | 0.99 | 0.992 | 9 | 63 | 500 | MyoD / ASC-related factors{1.2.2} | Myogenic transcription factors{1.2.2.1} | 7611 | 4654 | MYOD1_HUMAN | P15172 |
| MYOD1_HUMAN.H11MO.1.A |  |
HUMAN:MYOD1 | 11 | A | 1 | bnbCAGCTGTY | HOCOMOCOv11 | ChIP-Seq | 0.984 | 0.988 | 9 | 63 | 500 | MyoD / ASC-related factors{1.2.2} | Myogenic transcription factors{1.2.2.1} | 7611 | 4654 | MYOD1_HUMAN | P15172 |
| MYOG_HUMAN.H11MO.0.B |  |
HUMAN:MYOG | 13 | B | 0 | bhvCAGCTGYYhb | HOCOMOCOv11 | ChIP-Seq | 0.994 | 20 | 500 | MyoD / ASC-related factors{1.2.2} | Myogenic transcription factors{1.2.2.1} | 7612 | 4656 | MYOG_HUMAN | P15173 | ||
| MZF1_HUMAN.H11MO.0.B |  |
HUMAN:MZF1 | 13 | B | 0 | vvdRGGGAWbddv | HOCOMOCOv11 | ChIP-Seq | 0.851 | 6 | 503 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 13108 | 7593 | MZF1_HUMAN | P28698 | ||
| NANOG_HUMAN.H11MO.0.A |  |
HUMAN:NANOG | 17 | A | 0 | nbhWTTSWnATGYWRWb | HOCOMOCOv11 | ChIP-Seq | 0.92 | 0.928 | 15 | 38 | 499 | NK-related factors{3.1.2} | NANOG{3.1.2.12} | 20857 | 79923 | NANOG_HUMAN | Q9H9S0 |
| NANOG_HUMAN.H11MO.1.B |  |
HUMAN:NANOG | 11 | B | 1 | vRSCCATYAvn | HOCOMOCOv11 | ChIP-Seq | 0.769 | 0.825 | 15 | 38 | 500 | NK-related factors{3.1.2} | NANOG{3.1.2.12} | 20857 | 79923 | NANOG_HUMAN | Q9H9S0 |
| NDF1_HUMAN.H11MO.0.A |  |
HUMAN:NEUROD1 | 10 | A | 0 | RRCAGMTGGb | HOCOMOCOv11 | ChIP-Seq | 0.946 | 0.952 | 5 | 2 | 503 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 7762 | 4760 | NDF1_HUMAN | Q13562 |
| NDF2_HUMAN.H11MO.0.B |  |
HUMAN:NEUROD2 | 9 | B | 0 | RRCAGMTGG | HOCOMOCOv11 | ChIP-Seq | 0.594 | 0.963 | 2 | 21 | 502 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 7763 | 4761 | NDF2_HUMAN | Q15784 |
| NF2L1_HUMAN.H11MO.0.C |  |
HUMAN:NFE2L1 | 7 | C | 0 | hRTCAYn | HOCOMOCOv9 | Integrative | 115 | Jun-related factors{1.1.1} | NF-E2-like factors{1.1.1.2} | 7781 | 4779 | NF2L1_HUMAN | Q14494 | ||||
| NF2L2_HUMAN.H11MO.0.A |  |
HUMAN:NFE2L2 | 14 | A | 0 | WhhTGCTGAGTCAY | HOCOMOCOv11 | ChIP-Seq | 0.89 | 0.983 | 5 | 26 | 500 | Jun-related factors{1.1.1} | NF-E2-like factors{1.1.1.2} | 7782 | 4780 | NF2L2_HUMAN | Q16236 |
| NFAC1_HUMAN.H11MO.0.B |  |
HUMAN:NFATC1 | 15 | B | 0 | vWGGAAAvdvWRvvd | HOCOMOCOv11 | ChIP-Seq | 0.868 | 0.795 | 7 | 8 | 500 | NFAT-related factors{6.1.3} | NFATc1{6.1.3.0.1} | 7775 | 4772 | NFAC1_HUMAN | O95644 |
| NFAC1_HUMAN.H11MO.1.B |  |
HUMAN:NFATC1 | 10 | B | 1 | vvWGGAAARn | HOCOMOCOv11 | ChIP-Seq | 0.792 | 0.828 | 7 | 8 | 500 | NFAT-related factors{6.1.3} | NFATc1{6.1.3.0.1} | 7775 | 4772 | NFAC1_HUMAN | O95644 |
| NFAC2_HUMAN.H11MO.0.B |  |
HUMAN:NFATC2 | 9 | B | 0 | dGGAAAWhb | HOCOMOCOv9 | Integrative | 87 | NFAT-related factors{6.1.3} | NFATc2 (NFATp, NFAT1){6.1.3.0.2} | 7776 | 4773 | NFAC2_HUMAN | Q13469 | ||||
| NFAC3_HUMAN.H11MO.0.B |  |
HUMAN:NFATC3 | 9 | B | 0 | hGGAAAAhY | HOCOMOCOv9 | Integrative | 18 | NFAT-related factors{6.1.3} | NFATc3 (NFATx){6.1.3.0.3} | 7777 | 4775 | NFAC3_HUMAN | Q12968 | ||||
| NFAC4_HUMAN.H11MO.0.C |  |
HUMAN:NFATC4 | 10 | C | 0 | dGGAAARhdW | HOCOMOCOv9 | Integrative | 19 | NFAT-related factors{6.1.3} | NFATc4 (NFAT3){6.1.3.0.4} | 7778 | 4776 | NFAC4_HUMAN | Q14934 | ||||
| NFAT5_HUMAN.H11MO.0.D |  |
HUMAN:NFAT5 | 11 | D | 0 | vvdGGAARddn | HOCOMOCOv11 | ChIP-Seq | 0.706 | 3 | 500 | NFAT-related factors{6.1.3} | NFAT5 (TonEBP){6.1.3.0.5} | 7774 | 10725 | NFAT5_HUMAN | O94916 | ||
| NFE2_HUMAN.H11MO.0.A |  |
HUMAN:NFE2 | 13 | A | 0 | vvRTGACWCAGCW | HOCOMOCOv11 | ChIP-Seq | 0.937 | 0.991 | 17 | 9 | 500 | Jun-related factors{1.1.1} | NF-E2-like factors{1.1.1.2} | 7780 | 4778 | NFE2_HUMAN | Q16621 |
| NFIA_HUMAN.H11MO.0.C |  |
HUMAN:NFIA | 15 | C | 0 | hTGGShnGnhKCCWd | HOCOMOCOv9 | Integrative | 233 | Nuclear factor 1{7.1.2} | NF-1A (NF-IA){7.1.2.0.1} | 7784 | 4774 | NFIA_HUMAN | Q12857 | ||||
| NFIA_HUMAN.H11MO.1.D |  |
HUMAN:NFIA | 7 | D | 1 | hYTGGCd | HOCOMOCOv9 | Integrative | 267 | Nuclear factor 1{7.1.2} | NF-1A (NF-IA){7.1.2.0.1} | 7784 | 4774 | NFIA_HUMAN | Q12857 | ||||
| NFIB_HUMAN.H11MO.0.D |  |
HUMAN:NFIB | 9 | D | 0 | hYYTGGCWb | HOCOMOCOv11 | ChIP-Seq | 0.612 | 0.829 | 2 | 2 | 500 | Nuclear factor 1{7.1.2} | NF-1B (NF-IB){7.1.2.0.2} | 7785 | 4781 | NFIB_HUMAN | O00712 |
| NFIC_HUMAN.H11MO.0.A |  |
HUMAN:NFIC | 17 | A | 0 | bYKGGMhbChdKCCMdv | HOCOMOCOv11 | ChIP-Seq | 0.945 | 0.472 | 18 | 1 | 500 | Nuclear factor 1{7.1.2} | NF-1C (NF-IC){7.1.2.0.3} | 7786 | 4782 | NFIC_HUMAN | P08651 |
| NFIC_HUMAN.H11MO.1.A |  |
HUMAN:NFIC | 9 | A | 1 | bbTTGGCWb | HOCOMOCOv11 | ChIP-Seq | 0.932 | 0.831 | 18 | 1 | 500 | Nuclear factor 1{7.1.2} | NF-1C (NF-IC){7.1.2.0.3} | 7786 | 4782 | NFIC_HUMAN | P08651 |
| NFIL3_HUMAN.H11MO.0.D |  |
HUMAN:NFIL3 | 11 | D | 0 | dvTTATGYAAb | HOCOMOCOv11 | ChIP-Seq | 0.543 | 0.869 | 2 | 2 | 501 | C/EBP-related{1.1.8} | PAR factors{1.1.8.2} | 7787 | 4783 | NFIL3_HUMAN | Q16649 |
| NFKB1_HUMAN.H11MO.1.B |  |
HUMAN:NFKB1 | 11 | B | 1 | GGGAAAbYCCC | HOCOMOCOv11 | ChIP-Seq | 0.726 | 0.953 | 7 | 16 | 503 | NF-kappaB-related factors{6.1.1} | NF-kappaB p50 subunit-like factors{6.1.1.1} | 7794 | 4790 | NFKB1_HUMAN | P19838 |
| NFKB2_HUMAN.H11MO.0.B |  |
HUMAN:NFKB2 | 11 | B | 0 | KGGRAAdYCCC | HOCOMOCOv11 | ChIP-Seq | 0.977 | 4 | 500 | NF-kappaB-related factors{6.1.1} | NF-kappaB p50 subunit-like factors{6.1.1.1} | 7795 | 4791 | NFKB2_HUMAN | Q00653 | ||
| NFYA_HUMAN.H11MO.0.A |  |
HUMAN:NFYA | 14 | A | 0 | bbRRCCAATSRvMR | HOCOMOCOv11 | ChIP-Seq | 0.982 | 0.96 | 19 | 14 | 500 | Heteromeric CCAAT-binding factors{4.2.1} | NF-YA (CP1A, CBF-B){4.2.1.0.1} | 7804 | 4800 | NFYA_HUMAN | P23511 |
| NFYB_HUMAN.H11MO.0.A |  |
HUMAN:NFYB | 13 | A | 0 | bRRCCAATSRSvd | HOCOMOCOv9 | Integrative | 0.987 | 0.997 | 4 | 4 | 1163 | Heteromeric CCAAT-binding factors{4.2.1} | NF-YB (CP1B, CBF-A){4.2.1.0.2} | 7805 | 4801 | NFYB_HUMAN | P25208 |
| NFYC_HUMAN.H11MO.0.A |  |
HUMAN:NFYC | 14 | A | 0 | bbvdCCAATvRvvd | HOCOMOCOv11 | ChIP-Seq | 0.866 | 0.984 | 4 | 4 | 427 | Heteromeric CCAAT-binding factors{4.2.1} | NF-YC{4.2.1.0.3} | 7806 | 4802 | NFYC_HUMAN | Q13952 |
| NGN2_HUMAN.H11MO.0.D | 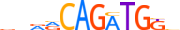 |
HUMAN:NEUROG2 | 12 | D | 0 | vbRRCAGATGKb | HOCOMOCOv11 | ChIP-Seq | 0.977 | 4 | 206 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 13805 | 63973 | NGN2_HUMAN | Q9H2A3 | ||
| NKX21_HUMAN.H11MO.0.A |  |
HUMAN:NKX2-1 | 10 | A | 0 | bWKGAGWGbh | HOCOMOCOv11 | ChIP-Seq | 0.92 | 0.917 | 14 | 9 | 501 | NK-related factors{3.1.2} | NK-2.1{3.1.2.14} | 11825 | 7080 | NKX21_HUMAN | P43699 |
| NKX22_HUMAN.H11MO.0.D |  |
HUMAN:NKX2-2 | 8 | D | 0 | dSAGWGvh | HOCOMOCOv11 | ChIP-Seq | 0.492 | 0.789 | 2 | 5 | 500 | NK-related factors{3.1.2} | NK-2.2{3.1.2.15} | 7835 | 4821 | NKX22_HUMAN | O95096 |
| NKX23_HUMAN.H11MO.0.D |  |
HUMAN:NKX2-3 | 11 | D | 0 | ndbAAGTGKbh | HOCOMOCOv10 | HT-SELEX | 1260 | NK-related factors{3.1.2} | NK-4{3.1.2.17} | 7836 | 159296 | NKX23_HUMAN | Q8TAU0 | ||||
| NKX25_HUMAN.H11MO.0.B |  |
HUMAN:NKX2-5 | 8 | B | 0 | bbRAGWGv | HOCOMOCOv11 | ChIP-Seq | 0.697 | 0.86 | 2 | 7 | 499 | NK-related factors{3.1.2} | NK-4{3.1.2.17} | 2488 | 1482 | NKX25_HUMAN | P52952 |
| NKX28_HUMAN.H11MO.0.C |  |
HUMAN:NKX2-8 | 9 | C | 0 | bTCAAGGAb | HOCOMOCOv9 | Integrative | 6 | NK-related factors{3.1.2} | NK-2.2{3.1.2.15} | 16364 | 26257 | NKX28_HUMAN | O15522 | ||||
| NKX31_HUMAN.H11MO.0.C |  |
HUMAN:NKX3-1 | 12 | C | 0 | nbdRAGTGbYWd | HOCOMOCOv11 | ChIP-Seq | 0.792 | 0.662 | 11 | 10 | 501 | NK-related factors{3.1.2} | NK-3{3.1.2.16} | 7838 | 4824 | NKX31_HUMAN | Q99801 |
| NKX32_HUMAN.H11MO.0.C |  |
HUMAN:NKX3-2 | 10 | C | 0 | WTAAGTGbhh | HOCOMOCOv11 | ChIP-Seq | 0.982 | 6 | 501 | NK-related factors{3.1.2} | NK-3{3.1.2.16} | 951 | 579 | NKX32_HUMAN | P78367 | ||
| NKX61_HUMAN.H11MO.0.B |  |
HUMAN:NKX6-1 | 19 | B | 0 | vRhdRATKdvndWWWRAWd | HOCOMOCOv11 | ChIP-Seq | 0.433 | 0.961 | 3 | 8 | 500 | NK-related factors{3.1.2} | NK-6{3.1.2.19} | 7839 | 4825 | NKX61_HUMAN | P78426 |
| NKX61_HUMAN.H11MO.1.B |  |
HUMAN:NKX6-1 | 7 | B | 1 | hAATGRv | HOCOMOCOv11 | ChIP-Seq | 0.547 | 0.846 | 3 | 8 | 500 | NK-related factors{3.1.2} | NK-6{3.1.2.19} | 7839 | 4825 | NKX61_HUMAN | P78426 |
| NKX62_HUMAN.H11MO.0.D |  |
HUMAN:NKX6-2 | 11 | D | 0 | ndWTAATKRbn | HOCOMOCOv10 | HT-SELEX | 8076 | NK-related factors{3.1.2} | NK-6{3.1.2.19} | 19321 | 84504 | NKX62_HUMAN | Q9C056 | ||||
| NOBOX_HUMAN.H11MO.0.C |  |
HUMAN:NOBOX | 9 | C | 0 | hTAATTGvb | HOCOMOCOv9 | Integrative | 66 | Paired-related HD factors{3.1.3} | NOBOX{3.1.3.15} | 22448 | 135935 | NOBOX_HUMAN | O60393 | ||||
| NOTO_HUMAN.H11MO.0.D |  |
HUMAN:NOTO | 11 | D | 0 | vYTAATTWRnb | HOCOMOCOv10 | HT-SELEX | 26150 | NK-related factors{3.1.2} | NOTO{3.1.2.20} | 31839 | 344022 | NOTO_HUMAN | A8MTQ0 | ||||
| NR0B1_HUMAN.H11MO.0.D |  |
HUMAN:NR0B1 | 10 | D | 0 | SCGKGGRASA | HOCOMOCOv9 | Integrative | 9 | DAX-related receptors (NR0){2.1.7} | DAX1 (NR0B1){2.1.7.0.1} | 7960 | 190 | NR0B1_HUMAN | P51843 | ||||
| NR1D1_HUMAN.H11MO.0.B |  |
HUMAN:NR1D1 | 19 | B | 0 | dRvnMWbWGKGdMAvdddv | HOCOMOCOv11 | ChIP-Seq | 0.615 | 0.883 | 1 | 53 | 501 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Rev-ErbA (NR1D){2.1.2.3} | 7962 | 9572 | NR1D1_HUMAN | P20393 |
| NR1D1_HUMAN.H11MO.1.D |  |
HUMAN:NR1D1 | 14 | D | 1 | WWAAvTAGGTCAnd | HOCOMOCOv9 | Integrative | 0.419 | 0.759 | 1 | 53 | 13 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Rev-ErbA (NR1D){2.1.2.3} | 7962 | 9572 | NR1D1_HUMAN | P20393 |
| NR1H2_HUMAN.H11MO.0.D |  |
HUMAN:NR1H2 | 19 | D | 0 | TMRAGGTCAAAGGTCAASK | HOCOMOCOv9 | Integrative | 24 | Thyroid hormone receptor-related factors (NR1){2.1.2} | LXR (NR1H){2.1.2.7} | 7965 | 7376 | NR1H2_HUMAN | P55055 | ||||
| NR1H3_HUMAN.H11MO.0.B |  |
HUMAN:NR1H3 | 19 | B | 0 | vhvRGKYhAbhnnRGKdYA | HOCOMOCOv11 | ChIP-Seq | 0.77 | 0.86 | 13 | 10 | 500 | Thyroid hormone receptor-related factors (NR1){2.1.2} | LXR (NR1H){2.1.2.7} | 7966 | 10062 | NR1H3_HUMAN | Q13133 |
| NR1H3_HUMAN.H11MO.1.B |  |
HUMAN:NR1H3 | 10 | B | 1 | hvAGGKCAvd | HOCOMOCOv11 | ChIP-Seq | 0.761 | 0.903 | 13 | 10 | 500 | Thyroid hormone receptor-related factors (NR1){2.1.2} | LXR (NR1H){2.1.2.7} | 7966 | 10062 | NR1H3_HUMAN | Q13133 |
| NR1H4_HUMAN.H11MO.0.B |  |
HUMAN:NR1H4 | 19 | B | 0 | vRGKKSRvYGdCCbbbvGK | HOCOMOCOv11 | ChIP-Seq | 0.926 | 15 | 451 | Thyroid hormone receptor-related factors (NR1){2.1.2} | LXR (NR1H){2.1.2.7} | 7967 | 9971 | NR1H4_HUMAN | Q96RI1 | ||
| NR1H4_HUMAN.H11MO.1.B |  |
HUMAN:NR1H4 | 8 | B | 1 | RGGKCAbh | HOCOMOCOv11 | ChIP-Seq | 0.849 | 15 | 501 | Thyroid hormone receptor-related factors (NR1){2.1.2} | LXR (NR1H){2.1.2.7} | 7967 | 9971 | NR1H4_HUMAN | Q96RI1 | ||
| NR1I2_HUMAN.H11MO.0.C |  |
HUMAN:NR1I2 | 19 | C | 0 | ddRRSTYARnnRAGKTCAb | HOCOMOCOv9 | Integrative | 29 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 7968 | 8856 | NR1I2_HUMAN | O75469 | ||||
| NR1I2_HUMAN.H11MO.1.D |  |
HUMAN:NR1I2 | 8 | D | 1 | dAGKTCAb | HOCOMOCOv9 | Integrative | 45 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 7968 | 8856 | NR1I2_HUMAN | O75469 | ||||
| NR1I3_HUMAN.H11MO.0.C |  |
HUMAN:NR1I3 | 18 | C | 0 | RRGSTCAvvvRAGKWYRd | HOCOMOCOv9 | Integrative | 28 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 7969 | 9970 | NR1I3_HUMAN | Q14994 | ||||
| NR1I3_HUMAN.H11MO.1.D |  |
HUMAN:NR1I3 | 8 | D | 1 | RAGTTCAb | HOCOMOCOv9 | Integrative | 33 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 7969 | 9970 | NR1I3_HUMAN | Q14994 | ||||
| NR2C1_HUMAN.H11MO.0.C |  |
HUMAN:NR2C1 | 13 | C | 0 | RbYvARAGGTCAn | HOCOMOCOv9 | Integrative | 19 | RXR-related receptors (NR2){2.1.3} | Testicular receptors (NR2C){2.1.3.4} | 7971 | 7181 | NR2C1_HUMAN | P13056 | ||||
| NR2C2_HUMAN.H11MO.0.B |  |
HUMAN:NR2C2 | 12 | B | 0 | RbTYMSGGGKCR | HOCOMOCOv11 | ChIP-Seq | 0.844 | 35 | 502 | RXR-related receptors (NR2){2.1.3} | Testicular receptors (NR2C){2.1.3.4} | 7972 | 7182 | NR2C2_HUMAN | P49116 | ||
| NR2E1_HUMAN.H11MO.0.D |  |
HUMAN:NR2E1 | 17 | D | 0 | MARKCAAdAAGTCAWbh | HOCOMOCOv10 | HT-SELEX | 70 | RXR-related receptors (NR2){2.1.3} | Tailless-like receptors (NR2E){2.1.3.3} | 7973 | 7101 | NR2E1_HUMAN | Q9Y466 | ||||
| NR2E3_HUMAN.H11MO.0.C |  |
HUMAN:NR2E3 | 14 | C | 0 | nAAKTCAAAARKMM | HOCOMOCOv9 | Integrative | 23 | RXR-related receptors (NR2){2.1.3} | Tailless-like receptors (NR2E){2.1.3.3} | 7974 | 10002 | NR2E3_HUMAN | Q9Y5X4 | ||||
| NR2F6_HUMAN.H11MO.0.D |  |
HUMAN:NR2F6 | 18 | D | 0 | RGSWMAAAGdnSAnnKdR | HOCOMOCOv9 | Integrative | 11 | RXR-related receptors (NR2){2.1.3} | COUP-like receptors (NR2F){2.1.3.5} | 7977 | 2063 | NR2F6_HUMAN | P10588 | ||||
| NR4A1_HUMAN.H11MO.0.A |  |
HUMAN:NR4A1 | 9 | A | 0 | AAAKKbYAn | HOCOMOCOv11 | ChIP-Seq | 0.92 | 0.547 | 12 | 2 | 491 | NGFI-B-related receptors (NR4){2.1.4} | NGFI-B (NR4A1){2.1.4.0.1} | 7980 | 3164 | NR4A1_HUMAN | P22736 |
| NR4A2_HUMAN.H11MO.0.C |  |
HUMAN:NR4A2 | 9 | C | 0 | RAAGGTCAv | HOCOMOCOv9 | Integrative | 33 | NGFI-B-related receptors (NR4){2.1.4} | NURR1 (NR4A2){2.1.4.0.2} | 7981 | 4929 | NR4A2_HUMAN | P43354 | ||||
| NR4A3_HUMAN.H11MO.0.D |  |
HUMAN:NR4A3 | 10 | D | 0 | CAAAGGTCAS | HOCOMOCOv9 | Integrative | 5 | NGFI-B-related receptors (NR4){2.1.4} | NOR1 (NR4A3){2.1.4.0.3} | 7982 | 8013 | NR4A3_HUMAN | Q92570 | ||||
| NR5A2_HUMAN.H11MO.0.B |  |
HUMAN:NR5A2 | 11 | B | 0 | dbYCAAGGhCR | HOCOMOCOv11 | ChIP-Seq | 0.555 | 0.959 | 4 | 8 | 500 | FTZ-F1-related receptors (NR5){2.1.5} | LRH-1 (NR5A2){2.1.5.0.2} | 7984 | 2494 | NR5A2_HUMAN | O00482 |
| NR6A1_HUMAN.H11MO.0.B |  |
HUMAN:NR6A1 | 13 | B | 0 | RAGKTCAAGKTCA | HOCOMOCOv9 | Integrative | 51 | GCNF-related receptors (NR6){2.1.6} | GCNF (NR6A1){2.1.6.0.1} | 7985 | 2649 | NR6A1_HUMAN | Q15406 | ||||
| NRF1_HUMAN.H11MO.0.A |  |
HUMAN:NRF1 | 17 | A | 0 | bdSYGCGCMKGCGCvdv | HOCOMOCOv11 | ChIP-Seq | 0.999 | 0.997 | 31 | 23 | 501 | NRF{0.0.6} | NRF-1 (alpha-pal){0.0.6.0.1} | 7996 | 4899 | NRF1_HUMAN | Q16656 |
| NRL_HUMAN.H11MO.0.D | 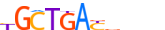 |
HUMAN:NRL | 11 | D | 0 | YGCTGAYdhdd | HOCOMOCOv10 | HT-SELEX | 805 | Maf-related factors{1.1.3} | Large Maf factors{1.1.3.1} | 8002 | 4901 | NRL_HUMAN | P54845 | ||||
| OLIG1_HUMAN.H11MO.0.D |  |
HUMAN:OLIG1 | 14 | D | 0 | AMCATATGTTTWbb | HOCOMOCOv10 | HT-SELEX | 495 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 16983 | 116448 | OLIG1_HUMAN | Q8TAK6 | ||||
| OLIG2_HUMAN.H11MO.0.B |  |
HUMAN:OLIG2 | 18 | B | 0 | MCAKCTGYYbbbhdbhhb | HOCOMOCOv11 | ChIP-Seq | 0.925 | 19 | 500 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 9398 | 10215 | OLIG2_HUMAN | Q13516 | ||
| OLIG2_HUMAN.H11MO.1.B |  |
HUMAN:OLIG2 | 10 | B | 1 | hMWKMTGYYb | HOCOMOCOv11 | ChIP-Seq | 0.905 | 19 | 500 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 9398 | 10215 | OLIG2_HUMAN | Q13516 | ||
| OLIG3_HUMAN.H11MO.0.D |  |
HUMAN:OLIG3 | 11 | D | 0 | RMCATATGKTn | HOCOMOCOv10 | HT-SELEX | 3197 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 18003 | 167826 | OLIG3_HUMAN | Q7RTU3 | ||||
| ONEC2_HUMAN.H11MO.0.D |  |
HUMAN:ONECUT2 | 21 | D | 0 | RKvhYdTbATKGATTWTWTTW | HOCOMOCOv9 | Integrative | 7 | HD-CUT factors{3.1.9} | ONECUT{3.1.9.1} | 8139 | 9480 | ONEC2_HUMAN | O95948 | ||||
| ONEC3_HUMAN.H11MO.0.D |  |
HUMAN:ONECUT3 | 12 | D | 0 | dWWbGATTTTTb | HOCOMOCOv10 | HT-SELEX | 10343 | HD-CUT factors{3.1.9} | ONECUT{3.1.9.1} | 13399 | 390874 | ONEC3_HUMAN | O60422 | ||||
| OSR2_HUMAN.H11MO.0.C |  |
HUMAN:OSR2 | 16 | C | 0 | nbhbYWGCTnCTShdb | HOCOMOCOv11 | ChIP-Seq | 0.898 | 3 | 492 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 15830 | 116039 | OSR2_HUMAN | Q8N2R0 | ||
| OTX1_HUMAN.H11MO.0.D |  |
HUMAN:OTX1 | 8 | D | 0 | nGGATTAn | HOCOMOCOv9 | Integrative | 8 | Paired-related HD factors{3.1.3} | OTX{3.1.3.17} | 8521 | 5013 | OTX1_HUMAN | P32242 | ||||
| OTX2_HUMAN.H11MO.0.A |  |
HUMAN:OTX2 | 11 | A | 0 | ddRGGATTAdv | HOCOMOCOv11 | ChIP-Seq | 0.863 | 0.886 | 5 | 18 | 500 | Paired-related HD factors{3.1.3} | OTX{3.1.3.17} | 8522 | 5015 | OTX2_HUMAN | P32243 |
| OVOL1_HUMAN.H11MO.0.C |  |
HUMAN:OVOL1 | 9 | C | 0 | KGTAACKGT | HOCOMOCOv9 | Integrative | 7 | More than 3 adjacent zinc finger factors{2.3.3} | OVOL-factors{2.3.3.17} | 8525 | 5017 | OVOL1_HUMAN | O14753 | ||||
| OVOL2_HUMAN.H11MO.0.D |  |
HUMAN:OVOL2 | 8 | D | 0 | dTAACGGd | HOCOMOCOv11 | ChIP-Seq | 0.453 | 0.952 | 2 | 2 | 197 | More than 3 adjacent zinc finger factors{2.3.3} | OVOL-factors{2.3.3.17} | 15804 | 58495 | OVOL2_HUMAN | Q9BRP0 |
| OZF_HUMAN.H11MO.0.C |  |
HUMAN:ZNF146 | 24 | C | 0 | TTTTYATGGCTGMATARTATTCCA | HOCOMOCOv11 | ChIP-Seq | 0.988 | 4 | 451 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF146-like factors{2.3.3.55} | 12931 | 7705 | OZF_HUMAN | Q15072 | ||
| P53_HUMAN.H11MO.0.A |  |
HUMAN:TP53 | 20 | A | 0 | dRRCWWGYYCWRRCATGYYh | HOCOMOCOv11 | ChIP-Seq | 0.995 | 0.979 | 141 | 39 | 487 | p53-related factors{6.3.1} | p53{6.3.1.0.1} | 11998 | 7157 | P53_HUMAN | P04637 |
| P53_HUMAN.H11MO.1.A |  |
HUMAN:TP53 | 13 | A | 1 | dRRCATGYCYRKR | HOCOMOCOv11 | ChIP-Seq | 0.988 | 0.981 | 141 | 39 | 500 | p53-related factors{6.3.1} | p53{6.3.1.0.1} | 11998 | 7157 | P53_HUMAN | P04637 |
| P5F1B_HUMAN.H11MO.0.D |  |
HUMAN:POU5F1B | 16 | D | 0 | dvWTTTGCATAWKYWd | HOCOMOCOv10 | HT-SELEX | 664 | POU domain factors{3.1.10} | POU5 (Oct-3/4-like factors){3.1.10.5} | 9223 | 5462 | P5F1B_HUMAN | Q06416 | ||||
| P63_HUMAN.H11MO.0.A |  |
HUMAN:TP63 | 19 | A | 0 | dRCRWGYhCWRRCWKGYhb | HOCOMOCOv11 | ChIP-Seq | 0.996 | 79 | 500 | p53-related factors{6.3.1} | p63{6.3.1.0.2} | 15979 | 8626 | P63_HUMAN | Q9H3D4 | ||
| P63_HUMAN.H11MO.1.A |  |
HUMAN:TP63 | 11 | A | 1 | vdRCMWGYCWR | HOCOMOCOv11 | ChIP-Seq | 0.936 | 79 | 502 | p53-related factors{6.3.1} | p63{6.3.1.0.2} | 15979 | 8626 | P63_HUMAN | Q9H3D4 | ||
| P73_HUMAN.H11MO.0.A |  |
HUMAN:TP73 | 19 | A | 0 | dddCWWGYYMMdRCWdGYh | HOCOMOCOv11 | ChIP-Seq | 0.996 | 10 | 500 | p53-related factors{6.3.1} | p73{6.3.1.0.3} | 12003 | 7161 | P73_HUMAN | O15350 | ||
| P73_HUMAN.H11MO.1.A |  |
HUMAN:TP73 | 12 | A | 1 | vdRCWWGYCYRR | HOCOMOCOv11 | ChIP-Seq | 0.908 | 10 | 500 | p53-related factors{6.3.1} | p73{6.3.1.0.3} | 12003 | 7161 | P73_HUMAN | O15350 | ||
| PATZ1_HUMAN.H11MO.0.C |  |
HUMAN:PATZ1 | 22 | C | 0 | dKRGvRRRGRMKSvRvndRvRb | HOCOMOCOv11 | ChIP-Seq | 0.917 | 6 | 498 | Factors with multiple dispersed zinc fingers{2.3.4} | MAZ-like factors{2.3.4.8} | 13071 | 23598 | PATZ1_HUMAN | Q9HBE1 | ||
| PATZ1_HUMAN.H11MO.1.C |  |
HUMAN:PATZ1 | 9 | C | 1 | dGGGAGRRG | HOCOMOCOv11 | ChIP-Seq | 0.859 | 6 | 499 | Factors with multiple dispersed zinc fingers{2.3.4} | MAZ-like factors{2.3.4.8} | 13071 | 23598 | PATZ1_HUMAN | Q9HBE1 | ||
| PAX1_HUMAN.H11MO.0.D |  |
HUMAN:PAX1 | 17 | D | 0 | hbGTCACGCWTSAnTKv | HOCOMOCOv10 | HT-SELEX | 1173 | Paired domain only{3.2.2} | PAX-1/9 (no homeo remnant){3.2.2.1} | 8615 | 5075 | PAX1_HUMAN | P15863 | ||||
| PAX2_HUMAN.H11MO.0.D |  |
HUMAN:PAX2 | 18 | D | 0 | vhbnvdTSAhRvdYGAYW | HOCOMOCOv9 | Integrative | 50 | Paired domain only{3.2.2} | PAX-2-like factors (partial homeobox){3.2.2.2} | 8616 | 5076 | PAX2_HUMAN | Q02962 | ||||
| PAX3_HUMAN.H11MO.0.D |  |
HUMAN:PAX3 | 13 | D | 0 | vhTAATCGATTAh | HOCOMOCOv10 | HT-SELEX | 5195 | Paired plus homeo domain{3.2.1} | PAX-3/7{3.2.1.1} | 8617 | 5077 | PAX3_HUMAN | P23760 | ||||
| PAX4_HUMAN.H11MO.0.D |  |
HUMAN:PAX4 | 12 | D | 0 | hdhhTAATKMdh | HOCOMOCOv10 | HT-SELEX | 934 | Paired plus homeo domain{3.2.1} | PAX-4/6{3.2.1.2} | 8618 | 5078 | PAX4_HUMAN | O43316 | ||||
| PAX5_HUMAN.H11MO.0.A |  |
HUMAN:PAX5 | 21 | A | 0 | vbvdGdKCARSvRRSCRdGAC | HOCOMOCOv11 | ChIP-Seq | 0.922 | 0.823 | 32 | 8 | 202 | Paired domain only{3.2.2} | PAX-2-like factors (partial homeobox){3.2.2.2} | 8619 | 5079 | PAX5_HUMAN | Q02548 |
| PAX6_HUMAN.H11MO.0.C | 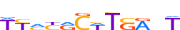 |
HUMAN:PAX6 | 12 | C | 0 | WYMYRCWTSRnT | HOCOMOCOv11 | ChIP-Seq | 0.749 | 0.734 | 6 | 8 | 500 | Paired plus homeo domain{3.2.1} | PAX-4/6{3.2.1.2} | 8620 | 5080 | PAX6_HUMAN | P26367 |
| PAX7_HUMAN.H11MO.0.D |  |
HUMAN:PAX7 | 11 | D | 0 | TAATYGATTAd | HOCOMOCOv10 | HT-SELEX | 17615 | Paired plus homeo domain{3.2.1} | PAX-3/7{3.2.1.1} | 8621 | 5081 | PAX7_HUMAN | P23759 | ||||
| PAX8_HUMAN.H11MO.0.D |  |
HUMAN:PAX8 | 14 | D | 0 | bYvAYYSRvSYRKd | HOCOMOCOv9 | Integrative | 37 | Paired domain only{3.2.2} | PAX-2-like factors (partial homeobox){3.2.2.2} | 8622 | 7849 | PAX8_HUMAN | Q06710 | ||||
| PBX1_HUMAN.H11MO.0.A |  |
HUMAN:PBX1 | 10 | A | 0 | dGAbKGACRR | HOCOMOCOv11 | ChIP-Seq | 0.936 | 0.883 | 2 | 36 | 501 | TALE-type homeo domain factors{3.1.4} | PBX{3.1.4.4} | 8632 | 5087 | PBX1_HUMAN | P40424 |
| PBX1_HUMAN.H11MO.1.C |  |
HUMAN:PBX1 | 15 | C | 1 | ddhWWGATKGATbdb | HOCOMOCOv9 | Integrative | 0.733 | 0.759 | 2 | 36 | 145 | TALE-type homeo domain factors{3.1.4} | PBX{3.1.4.4} | 8632 | 5087 | PBX1_HUMAN | P40424 |
| PBX2_HUMAN.H11MO.0.C |  |
HUMAN:PBX2 | 15 | C | 0 | KvMhKTGATTGWWKb | HOCOMOCOv9 | Integrative | 19 | TALE-type homeo domain factors{3.1.4} | PBX{3.1.4.4} | 8633 | 5089 | PBX2_HUMAN | P40425 | ||||
| PBX3_HUMAN.H11MO.0.A |  |
HUMAN:PBX3 | 11 | A | 0 | WGAbdGRCRKb | HOCOMOCOv11 | ChIP-Seq | 0.941 | 17 | 420 | TALE-type homeo domain factors{3.1.4} | PBX{3.1.4.4} | 8634 | 5090 | PBX3_HUMAN | P40426 | ||
| PBX3_HUMAN.H11MO.1.A |  |
HUMAN:PBX3 | 9 | A | 1 | YSAKTGGCh | HOCOMOCOv11 | ChIP-Seq | 0.922 | 17 | 500 | TALE-type homeo domain factors{3.1.4} | PBX{3.1.4.4} | 8634 | 5090 | PBX3_HUMAN | P40426 | ||
| PDX1_HUMAN.H11MO.0.A |  |
HUMAN:PDX1 | 9 | A | 0 | TGAYWRAKK | HOCOMOCOv11 | ChIP-Seq | 0.96 | 0.814 | 10 | 4 | 494 | HOX-related factors{3.1.1} | PDX{3.1.1.15} | 6107 | 3651 | PDX1_HUMAN | P52945 |
| PDX1_HUMAN.H11MO.1.A |  |
HUMAN:PDX1 | 9 | A | 1 | dYTAATKRb | HOCOMOCOv11 | ChIP-Seq | 0.824 | 0.827 | 10 | 4 | 504 | HOX-related factors{3.1.1} | PDX{3.1.1.15} | 6107 | 3651 | PDX1_HUMAN | P52945 |
| PEBB_HUMAN.H11MO.0.C |  |
HUMAN:CBFB | 11 | C | 0 | YbTGTGGTYdb | HOCOMOCOv9 | Integrative | 23 | 1539 | 865 | PEBB_HUMAN | Q13951 | ||||||
| PHX2A_HUMAN.H11MO.0.D |  |
HUMAN:PHOX2A | 11 | D | 0 | TAATYTAATTA | HOCOMOCOv10 | HT-SELEX | 7919 | Paired-related HD factors{3.1.3} | PHOX{3.1.3.18} | 691 | 401 | PHX2A_HUMAN | O14813 | ||||
| PHX2B_HUMAN.H11MO.0.D |  |
HUMAN:PHOX2B | 11 | D | 0 | WAATYTAATTA | HOCOMOCOv10 | HT-SELEX | 24552 | Paired-related HD factors{3.1.3} | PHOX{3.1.3.18} | 9143 | 8929 | PHX2B_HUMAN | Q99453 | ||||
| PIT1_HUMAN.H11MO.0.C |  |
HUMAN:POU1F1 | 13 | C | 0 | bWMWTGvAWATWh | HOCOMOCOv9 | Integrative | 39 | POU domain factors{3.1.10} | POU1 (Pit-1-like factors){3.1.10.1} | 9210 | 5449 | PIT1_HUMAN | P28069 | ||||
| PITX1_HUMAN.H11MO.0.D |  |
HUMAN:PITX1 | 9 | D | 0 | vRGGMTTWd | HOCOMOCOv11 | ChIP-Seq | 0.544 | 0.885 | 2 | 4 | 500 | Paired-related HD factors{3.1.3} | PITX{3.1.3.19} | 9004 | 5307 | PITX1_HUMAN | P78337 |
| PITX2_HUMAN.H11MO.0.D |  |
HUMAN:PITX2 | 10 | D | 0 | YKGGATTWvh | HOCOMOCOv9 | Integrative | 17 | Paired-related HD factors{3.1.3} | PITX{3.1.3.19} | 9005 | 5308 | PITX2_HUMAN | Q99697 | ||||
| PITX3_HUMAN.H11MO.0.D |  |
HUMAN:PITX3 | 18 | D | 0 | dddRdWWhvdvGGATTAd | HOCOMOCOv10 | HT-SELEX | 7586 | Paired-related HD factors{3.1.3} | PITX{3.1.3.19} | 9006 | 5309 | PITX3_HUMAN | O75364 | ||||
| PKNX1_HUMAN.H11MO.0.B |  |
HUMAN:PKNOX1 | 11 | B | 0 | WGAbKGRCRSv | HOCOMOCOv11 | ChIP-Seq | 0.967 | 19 | 480 | TALE-type homeo domain factors{3.1.4} | PKNOX{3.1.4.5} | 9022 | 5316 | PKNX1_HUMAN | P55347 | ||
| PLAG1_HUMAN.H11MO.0.D |  |
HUMAN:PLAG1 | 17 | D | 0 | RGGGGSMhndRdMGGGK | HOCOMOCOv9 | Integrative | 25 | More than 3 adjacent zinc finger factors{2.3.3} | PLAG factors{2.3.3.25} | 9045 | 5324 | PLAG1_HUMAN | Q6DJT9 | ||||
| PLAL1_HUMAN.H11MO.0.D |  |
HUMAN:PLAGL1 | 10 | D | 0 | vRGGGGGCCY | HOCOMOCOv9 | Integrative | 12 | More than 3 adjacent zinc finger factors{2.3.3} | PLAG factors{2.3.3.25} | 9046 | 5325 | PLAL1_HUMAN | Q9UM63 | ||||
| PO2F1_HUMAN.H11MO.0.C |  |
HUMAN:POU2F1 | 16 | C | 0 | RnKbvhATGSAAWhRh | HOCOMOCOv11 | ChIP-Seq | 0.766 | 0.669 | 15 | 14 | 134 | POU domain factors{3.1.10} | POU2 (Oct-1/2-like factors){3.1.10.2} | 9212 | 5451 | PO2F1_HUMAN | P14859 |
| PO2F2_HUMAN.H11MO.0.A |  |
HUMAN:POU2F2 | 11 | A | 0 | dhATGCAWAKd | HOCOMOCOv11 | ChIP-Seq | 0.919 | 14 | 501 | POU domain factors{3.1.10} | POU2 (Oct-1/2-like factors){3.1.10.2} | 9213 | 5452 | PO2F2_HUMAN | P09086 | ||
| PO2F3_HUMAN.H11MO.0.D |  |
HUMAN:POU2F3 | 14 | D | 0 | dWYAYGCWWRWdhd | HOCOMOCOv10 | HT-SELEX | 301 | POU domain factors{3.1.10} | POU2 (Oct-1/2-like factors){3.1.10.2} | 19864 | 25833 | PO2F3_HUMAN | Q9UKI9 | ||||
| PO3F1_HUMAN.H11MO.0.C |  |
HUMAN:POU3F1 | 14 | C | 0 | vRTKSTWATGCWWd | HOCOMOCOv9 | Integrative | 16 | POU domain factors{3.1.10} | POU3 (Oct-6-like factors){3.1.10.3} | 9214 | 5453 | PO3F1_HUMAN | Q03052 | ||||
| PO3F2_HUMAN.H11MO.0.A |  |
HUMAN:POU3F2 | 17 | A | 0 | YhCATKMMWWWbYMWdv | HOCOMOCOv11 | ChIP-Seq | 0.924 | 0.913 | 12 | 22 | 507 | POU domain factors{3.1.10} | POU3 (Oct-6-like factors){3.1.10.3} | 9215 | 5454 | PO3F2_HUMAN | P20265 |
| PO3F3_HUMAN.H11MO.0.D |  |
HUMAN:POU3F3 | 17 | D | 0 | hhWWWWWATKYWWWWdn | HOCOMOCOv10 | HT-SELEX | 1347 | POU domain factors{3.1.10} | POU3 (Oct-6-like factors){3.1.10.3} | 9216 | 5455 | PO3F3_HUMAN | P20264 | ||||
| PO3F4_HUMAN.H11MO.0.D |  |
HUMAN:POU3F4 | 17 | D | 0 | dhWTWWGCATAAWYhdd | HOCOMOCOv10 | HT-SELEX | 5283 | POU domain factors{3.1.10} | POU3 (Oct-6-like factors){3.1.10.3} | 9217 | 5456 | PO3F4_HUMAN | P49335 | ||||
| PO4F1_HUMAN.H11MO.0.D |  |
HUMAN:POU4F1 | 14 | D | 0 | hATWAATTATTYAd | HOCOMOCOv10 | HT-SELEX | 7624 | POU domain factors{3.1.10} | POU4 (Brn-3-like factors){3.1.10.4} | 9218 | 5457 | PO4F1_HUMAN | Q01851 | ||||
| PO4F2_HUMAN.H11MO.0.D |  |
HUMAN:POU4F2 | 13 | D | 0 | YvTTAATGAKhnn | HOCOMOCOv9 | Integrative | 31 | POU domain factors{3.1.10} | POU4 (Brn-3-like factors){3.1.10.4} | 9219 | 5458 | PO4F2_HUMAN | Q12837 | ||||
| PO4F3_HUMAN.H11MO.0.D |  |
HUMAN:POU4F3 | 15 | D | 0 | WhATTAATTATTYAh | HOCOMOCOv10 | HT-SELEX | 6312 | POU domain factors{3.1.10} | POU4 (Brn-3-like factors){3.1.10.4} | 9220 | 5459 | PO4F3_HUMAN | Q15319 | ||||
| PO5F1_HUMAN.H11MO.0.A | 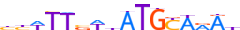 |
HUMAN:POU5F1 | 16 | A | 0 | bYWTTvWhATGYARAh | HOCOMOCOv11 | ChIP-Seq | 0.977 | 0.982 | 44 | 119 | 501 | POU domain factors{3.1.10} | POU5 (Oct-3/4-like factors){3.1.10.5} | 9221 | 5460 | PO5F1_HUMAN | Q01860 |
| PO5F1_HUMAN.H11MO.1.A |  |
HUMAN:POU5F1 | 10 | A | 1 | dnATGCAvWb | HOCOMOCOv11 | ChIP-Seq | 0.888 | 0.876 | 44 | 119 | 485 | POU domain factors{3.1.10} | POU5 (Oct-3/4-like factors){3.1.10.5} | 9221 | 5460 | PO5F1_HUMAN | Q01860 |
| PO6F1_HUMAN.H11MO.0.D |  |
HUMAN:POU6F1 | 13 | D | 0 | nATAATTTATGCR | HOCOMOCOv9 | Integrative | 14 | POU domain factors{3.1.10} | POU6 (Brn-5-like factors){3.1.10.6} | 9224 | 5463 | PO6F1_HUMAN | Q14863 | ||||
| PO6F2_HUMAN.H11MO.0.D |  |
HUMAN:POU6F2 | 11 | D | 0 | dhTAATTAGbh | HOCOMOCOv10 | HT-SELEX | 1481 | POU domain factors{3.1.10} | POU6 (Brn-5-like factors){3.1.10.6} | 21694 | 11281 | PO6F2_HUMAN | P78424 | ||||
| PPARA_HUMAN.H11MO.0.B |  |
HUMAN:PPARA | 17 | B | 0 | hWbKRGGbbARAGGKYR | HOCOMOCOv11 | ChIP-Seq | 0.576 | 0.956 | 5 | 20 | 500 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 9232 | 5465 | PPARA_HUMAN | Q07869 |
| PPARA_HUMAN.H11MO.1.B |  |
HUMAN:PPARA | 9 | B | 1 | MRAGGKCAd | HOCOMOCOv11 | ChIP-Seq | 0.553 | 0.833 | 5 | 20 | 500 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 9232 | 5465 | PPARA_HUMAN | Q07869 |
| PPARD_HUMAN.H11MO.0.D |  |
HUMAN:PPARD | 14 | D | 0 | YAGKnnAMAGGWYA | HOCOMOCOv9 | Integrative | 18 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 9235 | 5467 | PPARD_HUMAN | Q03181 | ||||
| PPARG_HUMAN.H11MO.0.A |  |
HUMAN:PPARG | 17 | A | 0 | vWbbRGGbSARAGGKSR | HOCOMOCOv11 | ChIP-Seq | 0.958 | 0.957 | 53 | 114 | 500 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 9236 | 5468 | PPARG_HUMAN | P37231 |
| PPARG_HUMAN.H11MO.1.A |  |
HUMAN:PPARG | 9 | A | 1 | SARAGKKhA | HOCOMOCOv11 | ChIP-Seq | 0.808 | 0.836 | 53 | 114 | 500 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 9236 | 5468 | PPARG_HUMAN | P37231 |
| PRD14_HUMAN.H11MO.0.A |  |
HUMAN:PRDM14 | 14 | A | 0 | ddSWTAGRGWMShn | HOCOMOCOv11 | ChIP-Seq | 0.914 | 0.889 | 7 | 11 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 14001 | 63978 | PRD14_HUMAN | Q9GZV8 |
| PRDM1_HUMAN.H11MO.0.A |  |
HUMAN:PRDM1 | 14 | A | 0 | RRRAGdGAAAGKdv | HOCOMOCOv11 | ChIP-Seq | 0.959 | 0.962 | 12 | 9 | 502 | More than 3 adjacent zinc finger factors{2.3.3} | PRDM1-like factors{2.3.3.12} | 9346 | 639 | PRDM1_HUMAN | O75626 |
| PRDM4_HUMAN.H11MO.0.D |  |
HUMAN:PRDM4 | 16 | D | 0 | bbGGGGRGMCTTGAAA | HOCOMOCOv10 | HT-SELEX | 130 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 9348 | 11108 | PRDM4_HUMAN | Q9UKN5 | ||||
| PRDM6_HUMAN.H11MO.0.C |  |
HUMAN:PRDM6 | 13 | C | 0 | vdvRRGAAAMMhv | HOCOMOCOv11 | ChIP-Seq | 0.951 | 2 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 9350 | 93166 | PRDM6_HUMAN | Q9NQX0 | ||
| PRGR_HUMAN.H11MO.0.A | 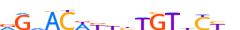 |
HUMAN:PGR | 16 | A | 0 | RGvACWYYbTGTdCYb | HOCOMOCOv11 | ChIP-Seq | 0.97 | 0.957 | 51 | 15 | 315 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 8910 | 5241 | PRGR_HUMAN | P06401 |
| PRGR_HUMAN.H11MO.1.A |  |
HUMAN:PGR | 9 | A | 1 | RGvACARvv | HOCOMOCOv11 | ChIP-Seq | 0.873 | 0.854 | 51 | 15 | 500 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 8910 | 5241 | PRGR_HUMAN | P06401 |
| PROP1_HUMAN.H11MO.0.D |  |
HUMAN:PROP1 | 11 | D | 0 | WAATKGvRTTW | HOCOMOCOv11 | ChIP-Seq | 0.921 | 3 | 500 | Paired-related HD factors{3.1.3} | PROP{3.1.3.20} | 9455 | 5626 | PROP1_HUMAN | O75360 | ||
| PROX1_HUMAN.H11MO.0.D |  |
HUMAN:PROX1 | 17 | D | 0 | bdbbRRGRhGKSdKRRd | HOCOMOCOv10 | HT-SELEX | 773 | HD-PROS factors{3.1.7} | PROX-1{3.1.7.0.1} | 9459 | 5629 | PROX1_HUMAN | Q92786 | ||||
| PRRX1_HUMAN.H11MO.0.D | 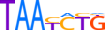 |
HUMAN:PRRX1 | 7 | D | 0 | TAATCTG | HOCOMOCOv9 | Integrative | 4 | Paired-related HD factors{3.1.3} | PRRX{3.1.3.21} | 9142 | 5396 | PRRX1_HUMAN | P54821 | ||||
| PRRX2_HUMAN.H11MO.0.C |  |
HUMAN:PRRX2 | 7 | C | 0 | hTAATTR | HOCOMOCOv9 | Integrative | 65 | Paired-related HD factors{3.1.3} | PRRX{3.1.3.21} | 21338 | 51450 | PRRX2_HUMAN | Q99811 | ||||
| PTF1A_HUMAN.H11MO.0.B |  |
HUMAN:PTF1A | 18 | B | 0 | vCAGCTGbbnbbbbYYhv | HOCOMOCOv11 | ChIP-Seq | 0.963 | 20 | 500 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 23734 | 256297 | PTF1A_HUMAN | Q7RTS3 | ||
| PTF1A_HUMAN.H11MO.1.B |  |
HUMAN:PTF1A | 9 | B | 1 | RRCAGMTGK | HOCOMOCOv11 | ChIP-Seq | 0.93 | 20 | 503 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 23734 | 256297 | PTF1A_HUMAN | Q7RTS3 | ||
| PURA_HUMAN.H11MO.0.D |  |
HUMAN:PURA | 17 | D | 0 | RGWvKbKGnKGSMRKGK | HOCOMOCOv9 | Integrative | 13 | PUR{0.0.5} | Pur-alpha (PURA, PUR1){0.0.5.0.1} | 9701 | 5813 | PURA_HUMAN | Q00577 | ||||
| RARA_HUMAN.H11MO.0.A |  |
HUMAN:RARA | 20 | A | 0 | vdddvdbbvdRAGKTCARRR | HOCOMOCOv11 | ChIP-Seq | 0.951 | 0.927 | 29 | 32 | 499 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 9864 | 5914 | RARA_HUMAN | P10276 |
| RARA_HUMAN.H11MO.1.A |  |
HUMAN:RARA | 10 | A | 1 | vAGGKCAvvv | HOCOMOCOv11 | ChIP-Seq | 0.948 | 0.874 | 29 | 32 | 501 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 9864 | 5914 | RARA_HUMAN | P10276 |
| RARA_HUMAN.H11MO.2.A |  |
HUMAN:RARA | 17 | A | 2 | RGKTSAnvvdvRSKKSR | HOCOMOCOv9 | Integrative | 0.806 | 0.844 | 29 | 32 | 122 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 9864 | 5914 | RARA_HUMAN | P10276 |
| RARB_HUMAN.H11MO.0.D |  |
HUMAN:RARB | 11 | D | 0 | vAGGTCAnvRv | HOCOMOCOv9 | Integrative | 25 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 9865 | 5915 | RARB_HUMAN | P10826 | ||||
| RARG_HUMAN.H11MO.0.B | 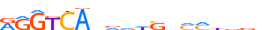 |
HUMAN:RARG | 18 | B | 0 | RRGTCAvvvhSvMMbddv | HOCOMOCOv11 | ChIP-Seq | 0.827 | 12 | 500 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 9866 | 5916 | RARG_HUMAN | P13631 | ||
| RARG_HUMAN.H11MO.1.B |  |
HUMAN:RARG | 8 | B | 1 | vAGGKCAn | HOCOMOCOv11 | ChIP-Seq | 0.815 | 12 | 394 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 9866 | 5916 | RARG_HUMAN | P13631 | ||
| RARG_HUMAN.H11MO.2.D |  |
HUMAN:RARG | 20 | D | 2 | dddGbdbRvMnvRAGGTCAv | HOCOMOCOv9 | Integrative | 0.656 | 12 | 24 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 9866 | 5916 | RARG_HUMAN | P13631 | ||
| RAX2_HUMAN.H11MO.0.D |  |
HUMAN:RAX2 | 12 | D | 0 | hTAATTRRhYhW | HOCOMOCOv10 | HT-SELEX | 33721 | Paired-related HD factors{3.1.3} | RAX{3.1.3.22} | 18286 | 84839 | RAX2_HUMAN | Q96IS3 | ||||
| RELB_HUMAN.H11MO.0.C |  |
HUMAN:RELB | 12 | C | 0 | vRGGGMTTTSMh | HOCOMOCOv9 | Integrative | 28 | NF-kappaB-related factors{6.1.1} | NF-kappaB p65 subunit-like factors{6.1.1.2} | 9956 | 5971 | RELB_HUMAN | Q01201 | ||||
| REL_HUMAN.H11MO.0.B |  |
HUMAN:REL | 15 | B | 0 | dvhdGGGRAWKbMSW | HOCOMOCOv11 | ChIP-Seq | 0.921 | 11 | 178 | NF-kappaB-related factors{6.1.1} | NF-kappaB p65 subunit-like factors{6.1.1.2} | 9954 | 5966 | REL_HUMAN | Q04864 | ||
| REST_HUMAN.H11MO.0.A |  |
HUMAN:REST | 22 | A | 0 | dTCAGSACCdhGGACAGMdShh | HOCOMOCOv11 | ChIP-Seq | 0.991 | 0.98 | 147 | 33 | 500 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 9966 | 5978 | REST_HUMAN | Q13127 |
| RFX1_HUMAN.H11MO.0.B |  |
HUMAN:RFX1 | 22 | B | 0 | bbbbSbGTTGCCWdGGvvMSvv | HOCOMOCOv10 | ChIP-Seq | 0.712 | 0.995 | 3 | 7 | 457 | RFX-related factors{3.3.3} | RFX1 (EF-C){3.3.3.0.1} | 9982 | 5989 | RFX1_HUMAN | P22670 |
| RFX1_HUMAN.H11MO.1.B | 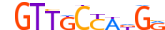 |
HUMAN:RFX1 | 11 | B | 1 | bGTTGCCWdGR | HOCOMOCOv11 | ChIP-Seq | 0.574 | 0.994 | 3 | 7 | 500 | RFX-related factors{3.3.3} | RFX1 (EF-C){3.3.3.0.1} | 9982 | 5989 | RFX1_HUMAN | P22670 |
| RFX2_HUMAN.H11MO.0.A |  |
HUMAN:RFX2 | 18 | A | 0 | YbGTTGCCWWGGvRACvv | HOCOMOCOv11 | ChIP-Seq | 0.976 | 0.991 | 7 | 7 | 426 | RFX-related factors{3.3.3} | RFX2{3.3.3.0.2} | 9983 | 5990 | RFX2_HUMAN | P48378 |
| RFX2_HUMAN.H11MO.1.A |  |
HUMAN:RFX2 | 12 | A | 1 | YbGTTGCCWKGG | HOCOMOCOv11 | ChIP-Seq | 0.965 | 0.996 | 7 | 7 | 501 | RFX-related factors{3.3.3} | RFX2{3.3.3.0.2} | 9983 | 5990 | RFX2_HUMAN | P48378 |
| RFX3_HUMAN.H11MO.0.B |  |
HUMAN:RFX3 | 14 | B | 0 | bGTTRCCATGGhRn | HOCOMOCOv9 | Integrative | 772 | RFX-related factors{3.3.3} | RFX3{3.3.3.0.3} | 9984 | 5991 | RFX3_HUMAN | P48380 | ||||
| RFX4_HUMAN.H11MO.0.D |  |
HUMAN:RFX4 | 15 | D | 0 | bGTTGCCATGGhWAY | HOCOMOCOv10 | HT-SELEX | 2791 | RFX-related factors{3.3.3} | RFX4{3.3.3.0.4} | 9985 | 5992 | RFX4_HUMAN | Q33E94 | ||||
| RFX5_HUMAN.H11MO.0.A |  |
HUMAN:RFX5 | 20 | A | 0 | vbbvbbdGTYRCYRKGbvvv | HOCOMOCOv11 | ChIP-Seq | 0.973 | 14 | 500 | RFX-related factors{3.3.3} | RFX5{3.3.3.0.5} | 9986 | 5993 | RFX5_HUMAN | P48382 | ||
| RFX5_HUMAN.H11MO.1.A |  |
HUMAN:RFX5 | 11 | A | 1 | YWGTTdCYRKG | HOCOMOCOv11 | ChIP-Seq | 0.926 | 14 | 167 | RFX-related factors{3.3.3} | RFX5{3.3.3.0.5} | 9986 | 5993 | RFX5_HUMAN | P48382 | ||
| RHXF1_HUMAN.H11MO.0.D |  |
HUMAN:RHOXF1 | 11 | D | 0 | nvbGGAThWdv | HOCOMOCOv10 | HT-SELEX | 3731 | Paired-related HD factors{3.1.3} | RHOX{3.1.3.23} | 29993 | 158800 | RHXF1_HUMAN | Q8NHV9 | ||||
| RORA_HUMAN.H11MO.0.C |  |
HUMAN:RORA | 13 | C | 0 | vhbdRGGbCAvdR | HOCOMOCOv11 | ChIP-Seq | 0.682 | 0.788 | 2 | 5 | 500 | Thyroid hormone receptor-related factors (NR1){2.1.2} | ROR (NR1F){2.1.2.6} | 10258 | 6095 | RORA_HUMAN | P35398 |
| RORG_HUMAN.H11MO.0.C |  |
HUMAN:RORC | 12 | C | 0 | vdRWSTRRGKCA | HOCOMOCOv11 | ChIP-Seq | 0.91 | 19 | 500 | Thyroid hormone receptor-related factors (NR1){2.1.2} | ROR (NR1F){2.1.2.6} | 10260 | 6097 | RORG_HUMAN | P51449 | ||
| RREB1_HUMAN.H11MO.0.D |  |
HUMAN:RREB1 | 22 | D | 0 | dbSKbdGKKGKTGbTKKGKGGd | HOCOMOCOv9 | Integrative | 26 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 10449 | 6239 | RREB1_HUMAN | Q92766 | ||||
| RUNX1_HUMAN.H11MO.0.A |  |
HUMAN:RUNX1 | 11 | A | 0 | bhTGTGGTYWn | HOCOMOCOv11 | ChIP-Seq | 0.964 | 0.983 | 115 | 63 | 500 | Runt-related factors{6.4.1} | Runx1 (PEBP2alphaB, CBF-alpha2, AML-1){6.4.1.0.2} | 10471 | 861 | RUNX1_HUMAN | Q01196 |
| RUNX2_HUMAN.H11MO.0.A |  |
HUMAN:RUNX2 | 14 | A | 0 | dbbbTGTGGYKKnb | HOCOMOCOv11 | ChIP-Seq | 0.965 | 0.894 | 3 | 25 | 502 | Runt-related factors{6.4.1} | Runx2 (PEBP2alphaA, CBF-alpha1){6.4.1.0.1} | 10472 | 860 | RUNX2_HUMAN | Q13950 |
| RUNX3_HUMAN.H11MO.0.A |  |
HUMAN:RUNX3 | 10 | A | 0 | bhTGTGGTYd | HOCOMOCOv11 | ChIP-Seq | 0.997 | 0.966 | 2 | 27 | 500 | Runt-related factors{6.4.1} | Runx3 (PEBP2alphaC, CBF-alpha3, AML-2){6.4.1.0.3} | 10473 | 864 | RUNX3_HUMAN | Q13761 |
| RXRA_HUMAN.H11MO.0.A |  |
HUMAN:RXRA | 20 | A | 0 | vdRdvddnvRRRGGKSAvdv | HOCOMOCOv11 | ChIP-Seq | 0.867 | 0.916 | 52 | 158 | 500 | RXR-related receptors (NR2){2.1.3} | Retinoid X receptors (NR2B){2.1.3.1} | 10477 | 6256 | RXRA_HUMAN | P19793 |
| RXRA_HUMAN.H11MO.1.A |  |
HUMAN:RXRA | 10 | A | 1 | vhRAGGTCAb | HOCOMOCOv11 | ChIP-Seq | 0.861 | 0.905 | 52 | 158 | 500 | RXR-related receptors (NR2){2.1.3} | Retinoid X receptors (NR2B){2.1.3.1} | 10477 | 6256 | RXRA_HUMAN | P19793 |
| RXRB_HUMAN.H11MO.0.C |  |
HUMAN:RXRB | 10 | C | 0 | WSAGGTCAbd | HOCOMOCOv9 | Integrative | 30 | RXR-related receptors (NR2){2.1.3} | Retinoid X receptors (NR2B){2.1.3.1} | 10478 | 6257 | RXRB_HUMAN | P28702 | ||||
| RXRG_HUMAN.H11MO.0.B |  |
HUMAN:RXRG | 13 | B | 0 | bGTCAAAGGTCAh | HOCOMOCOv9 | Integrative | 584 | RXR-related receptors (NR2){2.1.3} | Retinoid X receptors (NR2B){2.1.3.1} | 10479 | 6258 | RXRG_HUMAN | P48443 | ||||
| RX_HUMAN.H11MO.0.D |  |
HUMAN:RAX | 11 | D | 0 | hTAATTRvYhn | HOCOMOCOv10 | HT-SELEX | 340 | Paired-related HD factors{3.1.3} | RAX{3.1.3.22} | 18662 | 30062 | RX_HUMAN | Q9Y2V3 | ||||
| SALL4_HUMAN.H11MO.0.B |  |
HUMAN:SALL4 | 10 | B | 0 | dvdGGGWGKG | HOCOMOCOv11 | ChIP-Seq | 0.724 | 0.924 | 1 | 21 | 500 | Factors with multiple dispersed zinc fingers{2.3.4} | Sal-like factors{2.3.4.3} | 15924 | 57167 | SALL4_HUMAN | Q9UJQ4 |
| SCRT1_HUMAN.H11MO.0.D |  |
HUMAN:SCRT1 | 13 | D | 0 | KCAACAGGTKdbh | HOCOMOCOv10 | HT-SELEX | 179 | More than 3 adjacent zinc finger factors{2.3.3} | Snail-like factors{2.3.3.2} | 15950 | 83482 | SCRT1_HUMAN | Q9BWW7 | ||||
| SCRT2_HUMAN.H11MO.0.D |  |
HUMAN:SCRT2 | 13 | D | 0 | dhKCAACAGGTKb | HOCOMOCOv10 | HT-SELEX | 719 | More than 3 adjacent zinc finger factors{2.3.3} | Snail-like factors{2.3.3.2} | 15952 | 85508 | SCRT2_HUMAN | Q9NQ03 | ||||
| SHOX2_HUMAN.H11MO.0.D |  |
HUMAN:SHOX2 | 17 | D | 0 | bTAATTARbhhWWWWdd | HOCOMOCOv10 | HT-SELEX | 30757 | Paired-related HD factors{3.1.3} | SHOX{3.1.3.25} | 10854 | 6474 | SHOX2_HUMAN | O60902 | ||||
| SHOX_HUMAN.H11MO.0.D |  |
HUMAN:SHOX | 17 | D | 0 | bTAATTARbhhWWWWdd | HOCOMOCOv10 | HT-SELEX | 17808 | Paired-related HD factors{3.1.3} | SHOX{3.1.3.25} | 10853 | 6473 | SHOX_HUMAN | O15266 | ||||
| SIX1_HUMAN.H11MO.0.A | 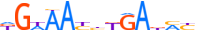 |
HUMAN:SIX1 | 14 | A | 0 | KGWAAYhKGAbMYb | HOCOMOCOv11 | ChIP-Seq | 0.961 | 6 | 500 | HD-SINE factors{3.1.6} | SIX1-like factors{3.1.6.1} | 10887 | 6495 | SIX1_HUMAN | Q15475 | ||
| SIX2_HUMAN.H11MO.0.A |  |
HUMAN:SIX2 | 13 | A | 0 | KGWAAYhhRAbMh | HOCOMOCOv11 | ChIP-Seq | 0.968 | 0.937 | 18 | 9 | 500 | HD-SINE factors{3.1.6} | SIX1-like factors{3.1.6.1} | 10888 | 10736 | SIX2_HUMAN | Q9NPC8 |
| SMAD1_HUMAN.H11MO.0.D | 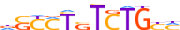 |
HUMAN:SMAD1 | 12 | D | 0 | dGCCTSTCTGYM | HOCOMOCOv9 | Integrative | 12 | SMAD factors{7.1.1} | Regulatory Smads (R-Smad){7.1.1.1} | 6767 | 4086 | SMAD1_HUMAN | Q15797 | ||||
| SMAD2_HUMAN.H11MO.0.A |  |
HUMAN:SMAD2 | 12 | A | 0 | WGKCTSnChCYb | HOCOMOCOv11 | ChIP-Seq | 0.848 | 0.808 | 17 | 26 | 500 | SMAD factors{7.1.1} | Regulatory Smads (R-Smad){7.1.1.1} | 6768 | 4087 | SMAD2_HUMAN | Q15796 |
| SMAD3_HUMAN.H11MO.0.B | 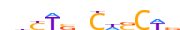 |
HUMAN:SMAD3 | 12 | B | 0 | vbYTShCWSCWb | HOCOMOCOv11 | ChIP-Seq | 0.806 | 0.707 | 34 | 30 | 500 | SMAD factors{7.1.1} | Regulatory Smads (R-Smad){7.1.1.1} | 6769 | 4088 | SMAD3_HUMAN | P84022 |
| SMAD4_HUMAN.H11MO.0.B |  |
HUMAN:SMAD4 | 13 | B | 0 | vWGKCTSdCWSYh | HOCOMOCOv11 | ChIP-Seq | 0.869 | 0.671 | 24 | 3 | 497 | SMAD factors{7.1.1} | Co-activating Smads (Co-Smad){7.1.1.2} | 6770 | 4089 | SMAD4_HUMAN | Q13485 |
| SMCA1_HUMAN.H11MO.0.C |  |
HUMAN:SMARCA1 | 12 | C | 0 | vMWRGAAKMAWn | HOCOMOCOv11 | ChIP-Seq | 0.912 | 3 | 442 | Myb/SANT domain factors{3.5.1} | SMARCA-like factors{3.5.1.5} | 11097 | 6594 | SMCA1_HUMAN | P28370 | ||
| SMCA5_HUMAN.H11MO.0.C |  |
HUMAN:SMARCA5 | 15 | C | 0 | GMvKnGAAWGSARWS | HOCOMOCOv11 | ChIP-Seq | 0.805 | 0.565 | 3 | 2 | 410 | Myb/SANT domain factors{3.5.1} | SMARCA-like factors{3.5.1.5} | 11101 | 8467 | SMCA5_HUMAN | O60264 |
| SNAI1_HUMAN.H11MO.0.C |  |
HUMAN:SNAI1 | 8 | C | 0 | vCAGGTGb | HOCOMOCOv9 | Integrative | 5 | More than 3 adjacent zinc finger factors{2.3.3} | Snail-like factors{2.3.3.2} | 11128 | 6615 | SNAI1_HUMAN | O95863 | ||||
| SNAI2_HUMAN.H11MO.0.A |  |
HUMAN:SNAI2 | 10 | A | 0 | dRCAGGTGYn | HOCOMOCOv11 | ChIP-Seq | 0.983 | 0.84 | 11 | 3 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | Snail-like factors{2.3.3.2} | 11094 | 6591 | SNAI2_HUMAN | O43623 |
| SOX10_HUMAN.H11MO.0.B | 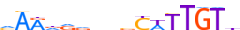 |
HUMAN:SOX10 | 16 | B | 0 | vARdRvvnbYWTTGTb | HOCOMOCOv11 | ChIP-Seq | 0.91 | 0.763 | 3 | 3 | 500 | SOX-related factors{4.1.1} | Group E{4.1.1.5} | 11190 | 6663 | SOX10_HUMAN | P56693 |
| SOX10_HUMAN.H11MO.1.A |  |
HUMAN:SOX10 | 13 | A | 1 | bbbCTTTSTbYhh | HOCOMOCOv11 | ChIP-Seq | 0.833 | 0.832 | 3 | 3 | 505 | SOX-related factors{4.1.1} | Group E{4.1.1.5} | 11190 | 6663 | SOX10_HUMAN | P56693 |
| SOX11_HUMAN.H11MO.0.D |  |
HUMAN:SOX11 | 16 | D | 0 | RACAMTRhMATTGTYb | HOCOMOCOv10 | HT-SELEX | 220 | SOX-related factors{4.1.1} | Group C{4.1.1.3} | 11191 | 6664 | SOX11_HUMAN | P35716 | ||||
| SOX13_HUMAN.H11MO.0.D |  |
HUMAN:SOX13 | 8 | D | 0 | nATTGTKn | HOCOMOCOv9 | Integrative | 5 | SOX-related factors{4.1.1} | Group D{4.1.1.4} | 11192 | 9580 | SOX13_HUMAN | Q9UN79 | ||||
| SOX15_HUMAN.H11MO.0.D |  |
HUMAN:SOX15 | 7 | D | 0 | CWTTGTT | HOCOMOCOv9 | Integrative | 8 | SOX-related factors{4.1.1} | Group G{4.1.1.7} | 11196 | 6665 | SOX15_HUMAN | O60248 | ||||
| SOX17_HUMAN.H11MO.0.C |  |
HUMAN:SOX17 | 11 | C | 0 | bCCATYKTbbb | HOCOMOCOv11 | ChIP-Seq | 0.835 | 4 | 500 | SOX-related factors{4.1.1} | Group F{4.1.1.6} | 18122 | 64321 | SOX17_HUMAN | Q9H6I2 | ||
| SOX18_HUMAN.H11MO.0.D |  |
HUMAN:SOX18 | 19 | D | 0 | SMWhnMATTGTYbWnnYYC | HOCOMOCOv9 | Integrative | 8 | SOX-related factors{4.1.1} | Group F{4.1.1.6} | 11194 | 54345 | SOX18_HUMAN | P35713 | ||||
| SOX1_HUMAN.H11MO.0.D |  |
HUMAN:SOX1 | 16 | D | 0 | dRTSAATGKTATTCAK | HOCOMOCOv10 | HT-SELEX | 40 | SOX-related factors{4.1.1} | Group B{4.1.1.2} | 11189 | 6656 | SOX1_HUMAN | O00570 | ||||
| SOX21_HUMAN.H11MO.0.D | 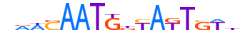 |
HUMAN:SOX21 | 16 | D | 0 | dRWSAATGdYAKTSWb | HOCOMOCOv10 | HT-SELEX | 14076 | SOX-related factors{4.1.1} | Group B{4.1.1.2} | 11197 | 11166 | SOX21_HUMAN | Q9Y651 | ||||
| SOX2_HUMAN.H11MO.0.A |  |
HUMAN:SOX2 | 13 | A | 0 | bbYCTTTGTbhbb | HOCOMOCOv11 | ChIP-Seq | 0.944 | 0.922 | 67 | 109 | 500 | SOX-related factors{4.1.1} | Group B{4.1.1.2} | 11195 | 6657 | SOX2_HUMAN | P48431 |
| SOX2_HUMAN.H11MO.1.A |  |
HUMAN:SOX2 | 16 | A | 1 | bYYWTTSTbhTKYhdW | HOCOMOCOv11 | ChIP-Seq | 0.957 | 0.955 | 67 | 109 | 500 | SOX-related factors{4.1.1} | Group B{4.1.1.2} | 11195 | 6657 | SOX2_HUMAN | P48431 |
| SOX3_HUMAN.H11MO.0.B |  |
HUMAN:SOX3 | 11 | B | 0 | bYCWTTGTbhb | HOCOMOCOv11 | ChIP-Seq | 0.717 | 0.882 | 2 | 4 | 500 | SOX-related factors{4.1.1} | Group B{4.1.1.2} | 11199 | 6658 | SOX3_HUMAN | P41225 |
| SOX4_HUMAN.H11MO.0.B |  |
HUMAN:SOX4 | 12 | B | 0 | bYCTTTGTYYbb | HOCOMOCOv11 | ChIP-Seq | 0.64 | 0.978 | 2 | 7 | 500 | SOX-related factors{4.1.1} | Group C{4.1.1.3} | 11200 | 6659 | SOX4_HUMAN | Q06945 |
| SOX5_HUMAN.H11MO.0.C |  |
HUMAN:SOX5 | 8 | C | 0 | bATTGTTh | HOCOMOCOv9 | Integrative | 69 | SOX-related factors{4.1.1} | Group D{4.1.1.4} | 11201 | 6660 | SOX5_HUMAN | P35711 | ||||
| SOX7_HUMAN.H11MO.0.D |  |
HUMAN:SOX7 | 16 | D | 0 | RACAMTGhMATTGTYh | HOCOMOCOv10 | HT-SELEX | 700 | SOX-related factors{4.1.1} | Group F{4.1.1.6} | 18196 | 83595 | SOX7_HUMAN | Q9BT81 | ||||
| SOX8_HUMAN.H11MO.0.D |  |
HUMAN:SOX8 | 16 | D | 0 | WvACTGhAATTSWYbh | HOCOMOCOv10 | HT-SELEX | 4026 | SOX-related factors{4.1.1} | Group E{4.1.1.5} | 11203 | 30812 | SOX8_HUMAN | P57073 | ||||
| SOX9_HUMAN.H11MO.0.B | 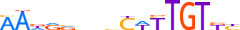 |
HUMAN:SOX9 | 16 | B | 0 | AAdRvnnbYWTTGTKY | HOCOMOCOv11 | ChIP-Seq | 0.718 | 0.981 | 6 | 25 | 500 | SOX-related factors{4.1.1} | Group E{4.1.1.5} | 11204 | 6662 | SOX9_HUMAN | P48436 |
| SOX9_HUMAN.H11MO.1.B |  |
HUMAN:SOX9 | 9 | B | 1 | YSWTTGKYY | HOCOMOCOv11 | ChIP-Seq | 0.777 | 0.892 | 6 | 25 | 501 | SOX-related factors{4.1.1} | Group E{4.1.1.5} | 11204 | 6662 | SOX9_HUMAN | P48436 |
| SP1_HUMAN.H11MO.0.A |  |
HUMAN:SP1 | 22 | A | 0 | nRGGGGCGGGGCSdSSvSSSvS | HOCOMOCOv11 | ChIP-Seq | 1.0 | 0.957 | 53 | 11 | 499 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 11205 | 6667 | SP1_HUMAN | P08047 |
| SP1_HUMAN.H11MO.1.A |  |
HUMAN:SP1 | 14 | A | 1 | vvKGGGCGGGGSbd | HOCOMOCOv11 | ChIP-Seq | 0.999 | 0.941 | 53 | 11 | 432 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 11205 | 6667 | SP1_HUMAN | P08047 |
| SP2_HUMAN.H11MO.0.A |  |
HUMAN:SP2 | 22 | A | 0 | SvvvvRRRGGCGGRRSbnvvSv | HOCOMOCOv10 | ChIP-Seq | 0.972 | 16 | 499 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 11207 | 6668 | SP2_HUMAN | Q02086 | ||
| SP2_HUMAN.H11MO.1.B |  |
HUMAN:SP2 | 12 | B | 1 | dRRGGGCGGGRM | HOCOMOCOv11 | ChIP-Seq | 0.893 | 16 | 498 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 11207 | 6668 | SP2_HUMAN | Q02086 | ||
| SP3_HUMAN.H11MO.0.B |  |
HUMAN:SP3 | 20 | B | 0 | vvvvvKGGGMGGRSvbdvvv | HOCOMOCOv9 | Integrative | 415 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 11208 | 6670 | SP3_HUMAN | Q02447 | ||||
| SP4_HUMAN.H11MO.0.A |  |
HUMAN:SP4 | 20 | A | 0 | vvSvdRRRGGMGGRGMhdvv | HOCOMOCOv11 | ChIP-Seq | 0.983 | 5 | 499 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 11209 | 6671 | SP4_HUMAN | Q02446 | ||
| SP4_HUMAN.H11MO.1.A |  |
HUMAN:SP4 | 16 | A | 1 | RvRRRGGGMGGRGvnd | HOCOMOCOv11 | ChIP-Seq | 0.983 | 5 | 496 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 11209 | 6671 | SP4_HUMAN | Q02446 | ||
| SPDEF_HUMAN.H11MO.0.D |  |
HUMAN:SPDEF | 14 | D | 0 | dWvCCGKATvWhvh | HOCOMOCOv10 | HT-SELEX | 1928 | Ets-related factors{3.5.2} | SPDEF-like factors{3.5.2.7} | 17257 | 25803 | SPDEF_HUMAN | O95238 | ||||
| SPI1_HUMAN.H11MO.0.A |  |
HUMAN:SPI1 | 17 | A | 0 | vvRRWRRGGAAGTGvvd | HOCOMOCOv11 | ChIP-Seq | 0.999 | 0.997 | 80 | 204 | 500 | Ets-related factors{3.5.2} | Spi-like factors{3.5.2.5} | 11241 | 6688 | SPI1_HUMAN | P17947 |
| SPIB_HUMAN.H11MO.0.A |  |
HUMAN:SPIB | 17 | A | 0 | vvRRWRRGGAAGTGRvd | HOCOMOCOv11 | ChIP-Seq | 0.998 | 0.949 | 12 | 3 | 500 | Ets-related factors{3.5.2} | Spi-like factors{3.5.2.5} | 11242 | 6689 | SPIB_HUMAN | Q01892 |
| SPIC_HUMAN.H11MO.0.D |  |
HUMAN:SPIC | 13 | D | 0 | dvddRWGKRdddd | HOCOMOCOv10 | HT-SELEX | 263 | Ets-related factors{3.5.2} | Spi-like factors{3.5.2.5} | 29549 | 121599 | SPIC_HUMAN | Q8N5J4 | ||||
| SPZ1_HUMAN.H11MO.0.D |  |
HUMAN:SPZ1 | 16 | D | 0 | nSRSKKTTMCMbhbGn | HOCOMOCOv9 | Integrative | 11 | 30721 | 84654 | SPZ1_HUMAN | Q9BXG8 | ||||||
| SRBP1_HUMAN.H11MO.0.A |  |
HUMAN:SREBF1 | 11 | A | 0 | dRKSRSGTGAb | HOCOMOCOv11 | ChIP-Seq | 0.92 | 0.67 | 5 | 23 | 500 | bHLH-ZIP factors{1.2.6} | SREBP factors{1.2.6.3} | 11289 | 6720 | SRBP1_HUMAN | P36956 |
| SRBP2_HUMAN.H11MO.0.B |  |
HUMAN:SREBF2 | 13 | B | 0 | vvvWGGvSWGRnb | HOCOMOCOv9 | Integrative | 2363 | bHLH-ZIP factors{1.2.6} | SREBP factors{1.2.6.3} | 11290 | 6721 | SRBP2_HUMAN | Q12772 | ||||
| SRF_HUMAN.H11MO.0.A |  |
HUMAN:SRF | 18 | A | 0 | hhbCCWWWTWTGGbvddd | HOCOMOCOv11 | ChIP-Seq | 0.956 | 0.979 | 46 | 37 | 131 | Responders to external signals (SRF/RLM1){5.1.2} | SRF{5.1.2.0.1} | 11291 | 6722 | SRF_HUMAN | P11831 |
| SRY_HUMAN.H11MO.0.B |  |
HUMAN:SRY | 9 | B | 0 | bWTTGTWWh | HOCOMOCOv9 | Integrative | 108 | SOX-related factors{4.1.1} | Group A{4.1.1.1} | 11311 | 6736 | SRY_HUMAN | Q05066 | ||||
| STA5A_HUMAN.H11MO.0.A |  |
HUMAN:STAT5A | 10 | A | 0 | TTCYARGAAd | HOCOMOCOv11 | ChIP-Seq | 0.932 | 0.992 | 24 | 127 | 500 | STAT factors{6.2.1} | STAT5A{6.2.1.0.5} | 11366 | 6776 | STA5A_HUMAN | P42229 |
| STA5B_HUMAN.H11MO.0.A |  |
HUMAN:STAT5B | 12 | A | 0 | nTTCYTRGAAdh | HOCOMOCOv11 | ChIP-Seq | 0.965 | 0.957 | 16 | 31 | 501 | STAT factors{6.2.1} | STAT5B{6.2.1.0.6} | 11367 | 6777 | STA5B_HUMAN | P51692 |
| STAT1_HUMAN.H11MO.0.A |  |
HUMAN:STAT1 | 19 | A | 0 | hbhnYTTCCnRGAARhShb | HOCOMOCOv11 | ChIP-Seq | 0.987 | 0.91 | 37 | 128 | 500 | STAT factors{6.2.1} | STAT1{6.2.1.0.1} | 11362 | 6772 | STAT1_HUMAN | P42224 |
| STAT1_HUMAN.H11MO.1.A |  |
HUMAN:STAT1 | 19 | A | 1 | vRRRAAhnGAAAShdRRdn | HOCOMOCOv11 | ChIP-Seq | 0.992 | 0.962 | 37 | 128 | 500 | STAT factors{6.2.1} | STAT1{6.2.1.0.1} | 11362 | 6772 | STAT1_HUMAN | P42224 |
| STAT2_HUMAN.H11MO.0.A |  |
HUMAN:STAT2 | 19 | A | 0 | dvRRAAhhGAAASYdRRRv | HOCOMOCOv11 | ChIP-Seq | 0.98 | 0.994 | 11 | 8 | 500 | STAT factors{6.2.1} | STAT2{6.2.1.0.2} | 11363 | 6773 | STAT2_HUMAN | P52630 |
| STAT3_HUMAN.H11MO.0.A |  |
HUMAN:STAT3 | 12 | A | 0 | bvhTTCCCRKAA | HOCOMOCOv11 | ChIP-Seq | 0.953 | 0.949 | 36 | 82 | 500 | STAT factors{6.2.1} | STAT3{6.2.1.0.3} | 11364 | 6774 | STAT3_HUMAN | P40763 |
| STAT4_HUMAN.H11MO.0.A | 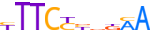 |
HUMAN:STAT4 | 11 | A | 0 | YTTCYYdKMAv | HOCOMOCOv11 | ChIP-Seq | 0.935 | 0.887 | 7 | 10 | 500 | STAT factors{6.2.1} | STAT4{6.2.1.0.4} | 11365 | 6775 | STAT4_HUMAN | Q14765 |
| STAT6_HUMAN.H11MO.0.B |  |
HUMAN:STAT6 | 11 | B | 0 | TTCYbWRGAAv | HOCOMOCOv11 | ChIP-Seq | 0.766 | 0.912 | 19 | 24 | 502 | STAT factors{6.2.1} | STAT6{6.2.1.0.7} | 11368 | 6778 | STAT6_HUMAN | P42226 |
| STF1_HUMAN.H11MO.0.B |  |
HUMAN:NR5A1 | 11 | B | 0 | bbYCAAGGYCW | HOCOMOCOv11 | ChIP-Seq | 0.759 | 0.898 | 1 | 2 | 500 | FTZ-F1-related receptors (NR5){2.1.5} | FTZ-F1 (SF-1) (NR5A1){2.1.5.0.1} | 7983 | 2516 | STF1_HUMAN | Q13285 |
| SUH_HUMAN.H11MO.0.A |  |
HUMAN:RBPJ | 11 | A | 0 | vbbSWGRGAAv | HOCOMOCOv11 | ChIP-Seq | 0.925 | 0.973 | 25 | 6 | 500 | CSL-related factors{6.1.4} | M{6.1.4.1} | 5724 | 3516 | SUH_HUMAN | Q06330 |
| TAF1_HUMAN.H11MO.0.A |  |
HUMAN:TAF1 | 16 | A | 0 | RRRRhGGMRGhRSvvv | HOCOMOCOv11 | ChIP-Seq | 0.894 | 0.91 | 61 | 2 | 500 | TCF-7-related factors{4.1.3} | TAF-1 (TAF-2A, TAF(II)250) [1]{4.1.3.0.5} | 11535 | 6872 | TAF1_HUMAN | P21675 |
| TAL1_HUMAN.H11MO.0.A |  |
HUMAN:TAL1 | 19 | A | 0 | bKKbnnnnnvvWGATAASv | HOCOMOCOv11 | ChIP-Seq | 0.978 | 0.968 | 68 | 76 | 500 | Tal-related factors{1.2.3} | Tal / HEN-like factors{1.2.3.1} | 11556 | 6886 | TAL1_HUMAN | P17542 |
| TAL1_HUMAN.H11MO.1.A |  |
HUMAN:TAL1 | 15 | A | 1 | vvdvhRRCAGvTGbb | HOCOMOCOv11 | ChIP-Seq | 0.828 | 0.73 | 68 | 76 | 500 | Tal-related factors{1.2.3} | Tal / HEN-like factors{1.2.3.1} | 11556 | 6886 | TAL1_HUMAN | P17542 |
| TBP_HUMAN.H11MO.0.A |  |
HUMAN:TBP | 10 | A | 0 | MbATAWAWRS | HOCOMOCOv11 | ChIP-Seq | 0.868 | 0.798 | 21 | 45 | 149 | TBP-related factors{8.1.1} | TBP{8.1.1.0.1} | 11588 | 6908 | TBP_HUMAN | P20226 |
| TBR1_HUMAN.H11MO.0.D |  |
HUMAN:TBR1 | 13 | D | 0 | vdAGGTGTGAWhd | HOCOMOCOv10 | HT-SELEX | 16871 | TBrain-related factors{6.5.2} | TBR-1{6.5.2.0.1} | 11590 | 10716 | TBR1_HUMAN | Q16650 | ||||
| TBX15_HUMAN.H11MO.0.D |  |
HUMAN:TBX15 | 19 | D | 0 | KKKKdKKKddRGGYGGGdd | HOCOMOCOv10 | HT-SELEX | 6956 | TBX1-related factors{6.5.3} | TBX15 (TBX14){6.5.3.0.3} | 11594 | 6913 | TBX15_HUMAN | Q96SF7 | ||||
| TBX19_HUMAN.H11MO.0.D |  |
HUMAN:TBX19 | 22 | D | 0 | dddTMRCAYnTAKGTGTKAhhd | HOCOMOCOv10 | HT-SELEX | 16587 | Brachyury-related factors{6.5.1} | TBX19{6.5.1.0.2} | 11596 | 9095 | TBX19_HUMAN | O60806 | ||||
| TBX1_HUMAN.H11MO.0.D |  |
HUMAN:TBX1 | 20 | D | 0 | RKKbGGGhRGGGGYGGGhdd | HOCOMOCOv10 | HT-SELEX | 4716 | TBX1-related factors{6.5.3} | TBX1{6.5.3.0.1} | 11592 | 6899 | TBX1_HUMAN | O43435 | ||||
| TBX20_HUMAN.H11MO.0.D |  |
HUMAN:TBX20 | 14 | D | 0 | ddSTGnTGAMARvn | HOCOMOCOv11 | ChIP-Seq | 0.883 | 4 | 500 | TBX1-related factors{6.5.3} | TBX20{6.5.3.0.5} | 11598 | 57057 | TBX20_HUMAN | Q9UMR3 | ||
| TBX21_HUMAN.H11MO.0.A |  |
HUMAN:TBX21 | 13 | A | 0 | vdvddRSTGWKRn | HOCOMOCOv11 | ChIP-Seq | 0.829 | 0.831 | 7 | 17 | 497 | TBrain-related factors{6.5.2} | TBX21 (T-bet){6.5.2.0.3} | 11599 | 30009 | TBX21_HUMAN | Q9UL17 |
| TBX2_HUMAN.H11MO.0.D |  |
HUMAN:TBX2 | 25 | D | 0 | hbMhWhChbAGGTGTGAnRWKnSMn | HOCOMOCOv9 | Integrative | 9 | TBX2-related factors{6.5.4} | TBX2{6.5.4.0.1} | 11597 | 6909 | TBX2_HUMAN | Q13207 | ||||
| TBX3_HUMAN.H11MO.0.C |  |
HUMAN:TBX3 | 11 | C | 0 | dvWGGTGbnRv | HOCOMOCOv11 | ChIP-Seq | 0.963 | 0.583 | 2 | 3 | 420 | TBX2-related factors{6.5.4} | TBX3{6.5.4.0.2} | 11602 | 6926 | TBX3_HUMAN | O15119 |
| TBX4_HUMAN.H11MO.0.D | 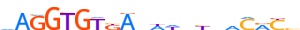 |
HUMAN:TBX4 | 20 | D | 0 | dAGGTGYKAhWhnYvMMMYb | HOCOMOCOv10 | HT-SELEX | 16745 | TBX2-related factors{6.5.4} | TBX4{6.5.4.0.3} | 11603 | 9496 | TBX4_HUMAN | P57082 | ||||
| TBX5_HUMAN.H11MO.0.D |  |
HUMAN:TBX5 | 8 | D | 0 | ARGTGKSW | HOCOMOCOv9 | Integrative | 38 | TBX2-related factors{6.5.4} | TBX5{6.5.4.0.4} | 11604 | 6910 | TBX5_HUMAN | Q99593 | ||||
| TCF7_HUMAN.H11MO.0.A |  |
HUMAN:TCF7 | 16 | A | 0 | dvhvWSWTSAAARvnv | HOCOMOCOv11 | ChIP-Seq | 0.847 | 0.929 | 7 | 19 | 500 | TCF-7-related factors{4.1.3} | TCF-7 (TCF-1) [1]{4.1.3.0.1} | 11639 | 6932 | TCF7_HUMAN | P36402 |
| TEAD1_HUMAN.H11MO.0.A |  |
HUMAN:TEAD1 | 12 | A | 0 | RCATTCCWdvvh | HOCOMOCOv11 | ChIP-Seq | 0.969 | 0.93 | 14 | 5 | 500 | TEF-1-related factors{3.6.1} | TEF-1 (TEAD-1, TCF-13){3.6.1.0.1} | 11714 | 7003 | TEAD1_HUMAN | P28347 |
| TEAD2_HUMAN.H11MO.0.D |  |
HUMAN:TEAD2 | 12 | D | 0 | hbRMATTCCWRv | HOCOMOCOv11 | ChIP-Seq | 0.605 | 0.919 | 2 | 2 | 308 | TEF-1-related factors{3.6.1} | TEF-4 (TEAD-2){3.6.1.0.3} | 11715 | 8463 | TEAD2_HUMAN | Q15562 |
| TEAD3_HUMAN.H11MO.0.D |  |
HUMAN:TEAD3 | 15 | D | 0 | nAWATTYCTGhhbYh | HOCOMOCOv9 | Integrative | 12 | TEF-1-related factors{3.6.1} | TEF-5 (TEAD-3, TEAD5){3.6.1.0.4} | 11716 | 7005 | TEAD3_HUMAN | Q99594 | ||||
| TEAD4_HUMAN.H11MO.0.A |  |
HUMAN:TEAD4 | 13 | A | 0 | RMATTCCWdvvhb | HOCOMOCOv11 | ChIP-Seq | 0.971 | 0.935 | 57 | 10 | 500 | TEF-1-related factors{3.6.1} | TEF-3 (TEAD-4, TCF-13L1){3.6.1.0.2} | 11717 | 7004 | TEAD4_HUMAN | Q15561 |
| TEF_HUMAN.H11MO.0.D |  |
HUMAN:TEF | 14 | D | 0 | TGdTKAYRTMdvYd | HOCOMOCOv9 | Integrative | 14 | C/EBP-related{1.1.8} | PAR factors{1.1.8.2} | 11722 | 7008 | TEF_HUMAN | Q10587 | ||||
| TF2LX_HUMAN.H11MO.0.D |  |
HUMAN:TGIF2LX | 15 | D | 0 | RYhnTdACAKCTnTC | HOCOMOCOv10 | HT-SELEX | 14 | TALE-type homeo domain factors{3.1.4} | TGIF{3.1.4.6} | 18570 | 90316 | TF2LX_HUMAN | Q8IUE1 | ||||
| TF65_HUMAN.H11MO.0.A |  |
HUMAN:RELA | 14 | A | 0 | hdGGGRhTTTCChv | HOCOMOCOv11 | ChIP-Seq | 0.977 | 0.906 | 188 | 99 | 500 | NF-kappaB-related factors{6.1.1} | NF-kappaB p65 subunit-like factors{6.1.1.2} | 9955 | 5970 | TF65_HUMAN | Q04206 |
| TF7L1_HUMAN.H11MO.0.B |  |
HUMAN:TCF7L1 | 13 | B | 0 | dbbCTTTSAWSWb | HOCOMOCOv11 | ChIP-Seq | 0.926 | 0.741 | 10 | 10 | 500 | TCF-7-related factors{4.1.3} | TCF-7L1 (TCF-3) [1]{4.1.3.0.2} | 11640 | 83439 | TF7L1_HUMAN | Q9HCS4 |
| TF7L2_HUMAN.H11MO.0.A |  |
HUMAN:TCF7L2 | 13 | A | 0 | bbCTTTGATSYbb | HOCOMOCOv11 | ChIP-Seq | 0.962 | 0.888 | 46 | 10 | 500 | TCF-7-related factors{4.1.3} | TCF-7L2 (TCF-4) [1]{4.1.3.0.3} | 11641 | 6934 | TF7L2_HUMAN | Q9NQB0 |
| TFAP4_HUMAN.H11MO.0.A |  |
HUMAN:TFAP4 | 10 | A | 0 | hCAGCTGdKb | HOCOMOCOv11 | ChIP-Seq | 0.923 | 7 | 503 | bHLH-ZIP factors{1.2.6} | AP-4{1.2.6.4} | 11745 | 7023 | TFAP4_HUMAN | Q01664 | ||
| TFCP2_HUMAN.H11MO.0.D |  |
HUMAN:TFCP2 | 16 | D | 0 | SSCWGvnMdvRCMRGM | HOCOMOCOv9 | Integrative | 13 | CP2-related factors{6.7.2} | CP2 (LSF, SEF){6.7.2.0.1} | 11748 | 7024 | TFCP2_HUMAN | Q12800 | ||||
| TFDP1_HUMAN.H11MO.0.C |  |
HUMAN:TFDP1 | 14 | C | 0 | vvddSGCGGGAMdn | HOCOMOCOv11 | ChIP-Seq | 0.871 | 3 | 500 | E2F-related factors{3.3.2} | Dp-1{3.3.2.2} | 11749 | 7027 | TFDP1_HUMAN | Q14186 | ||
| TFE2_HUMAN.H11MO.0.A |  |
HUMAN:TCF3 | 11 | A | 0 | vMCASCTGShn | HOCOMOCOv11 | ChIP-Seq | 0.926 | 0.977 | 9 | 60 | 503 | E2A-related factors{1.2.1} | E2A (TCF-3, ITF-1){1.2.1.0.1} | 11633 | 6929 | TFE2_HUMAN | P15923 |
| TFE3_HUMAN.H11MO.0.B |  |
HUMAN:TFE3 | 10 | B | 0 | dGTCACGTGA | HOCOMOCOv11 | ChIP-Seq | 0.483 | 1.0 | 2 | 30 | 478 | bHLH-ZIP factors{1.2.6} | TFE3-like factors{1.2.6.1} | 11752 | 7030 | TFE3_HUMAN | P19532 |
| TFEB_HUMAN.H11MO.0.C | 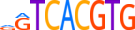 |
HUMAN:TFEB | 9 | C | 0 | dGTCACGTG | HOCOMOCOv9 | Integrative | 301 | bHLH-ZIP factors{1.2.6} | TFE3-like factors{1.2.6.1} | 11753 | 7942 | TFEB_HUMAN | P19484 | ||||
| TGIF1_HUMAN.H11MO.0.A |  |
HUMAN:TGIF1 | 7 | A | 0 | WGACAGb | HOCOMOCOv11 | ChIP-Seq | 0.861 | 0.976 | 2 | 5 | 500 | TALE-type homeo domain factors{3.1.4} | TGIF{3.1.4.6} | 11776 | 7050 | TGIF1_HUMAN | Q15583 |
| TGIF2_HUMAN.H11MO.0.D |  |
HUMAN:TGIF2 | 11 | D | 0 | nTGACASvYRY | HOCOMOCOv10 | HT-SELEX | 46893 | TALE-type homeo domain factors{3.1.4} | TGIF{3.1.4.6} | 15764 | 60436 | TGIF2_HUMAN | Q9GZN2 | ||||
| THA11_HUMAN.H11MO.0.B |  |
HUMAN:THAP11 | 22 | B | 0 | KGSMTbCTGGGAvWTGTAGTYY | HOCOMOCOv11 | ChIP-Seq | 0.942 | 0.768 | 2 | 2 | 126 | THAP-related factors{2.9.1} | THAP11 (HRIHFB2206){2.9.1.0.11} | 23194 | 57215 | THA11_HUMAN | Q96EK4 |
| THAP1_HUMAN.H11MO.0.C |  |
HUMAN:THAP1 | 22 | C | 0 | vSSvKCCMYbKYbvnbbhKGSY | HOCOMOCOv10 | ChIP-Seq | 0.977 | 4 | 498 | THAP-related factors{2.9.1} | THAP1{2.9.1.0.1} | 20856 | 55145 | THAP1_HUMAN | Q9NVV9 | ||
| THA_HUMAN.H11MO.0.C | 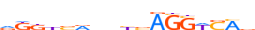 |
HUMAN:THRA | 17 | C | 0 | dKKhvRnhbvAGGWSMb | HOCOMOCOv9 | Integrative | 75 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Thyroid hormone receptors (NR1A){2.1.2.2} | 11796 | 7067 | THA_HUMAN | P10827 | ||||
| THA_HUMAN.H11MO.1.D |  |
HUMAN:THRA | 10 | D | 1 | nYvAGGTCAv | HOCOMOCOv9 | Integrative | 76 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Thyroid hormone receptors (NR1A){2.1.2.2} | 11796 | 7067 | THA_HUMAN | P10827 | ||||
| THB_HUMAN.H11MO.0.C | 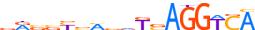 |
HUMAN:THRB | 17 | C | 0 | vRvSYvMbvKSAGGTCA | HOCOMOCOv9 | Integrative | 38 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Thyroid hormone receptors (NR1A){2.1.2.2} | 11799 | 7068 | THB_HUMAN | P10828 | ||||
| THB_HUMAN.H11MO.1.D |  |
HUMAN:THRB | 10 | D | 1 | bvTSAGGTCA | HOCOMOCOv9 | Integrative | 58 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Thyroid hormone receptors (NR1A){2.1.2.2} | 11799 | 7068 | THB_HUMAN | P10828 | ||||
| TLX1_HUMAN.H11MO.0.D |  |
HUMAN:TLX1 | 17 | D | 0 | hYKKYAvbndvMYnRKd | HOCOMOCOv9 | Integrative | 50 | NK-related factors{3.1.2} | TLX{3.1.2.21} | 5056 | 3195 | TLX1_HUMAN | P31314 | ||||
| TWST1_HUMAN.H11MO.0.A |  |
HUMAN:TWIST1 | 17 | A | 0 | vCAKMTGKWWbbvRTYW | HOCOMOCOv11 | ChIP-Seq | 0.969 | 0.539 | 9 | 8 | 497 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 12428 | 7291 | TWST1_HUMAN | Q15672 |
| TWST1_HUMAN.H11MO.1.A |  |
HUMAN:TWIST1 | 10 | A | 1 | nvCATSTGKW | HOCOMOCOv11 | ChIP-Seq | 0.971 | 0.567 | 9 | 8 | 502 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 12428 | 7291 | TWST1_HUMAN | Q15672 |
| TYY1_HUMAN.H11MO.0.A |  |
HUMAN:YY1 | 12 | A | 0 | MAARATGGCKSM | HOCOMOCOv11 | ChIP-Seq | 0.981 | 0.989 | 69 | 72 | 453 | More than 3 adjacent zinc finger factors{2.3.3} | YY1-like factors{2.3.3.9} | 12856 | 7528 | TYY1_HUMAN | P25490 |
| TYY2_HUMAN.H11MO.0.D |  |
HUMAN:YY2 | 11 | D | 0 | WhvATGGCGGn | HOCOMOCOv10 | HT-SELEX | 943 | More than 3 adjacent zinc finger factors{2.3.3} | YY1-like factors{2.3.3.9} | 31684 | 404281 | TYY2_HUMAN | O15391 | ||||
| UBIP1_HUMAN.H11MO.0.D |  |
HUMAN:UBP1 | 7 | D | 0 | nCAGAdA | HOCOMOCOv9 | Integrative | 5 | CP2-related factors{6.7.2} | UBP-1 (LBP-1, SEF){6.7.2.0.3} | 12507 | 7342 | UBIP1_HUMAN | Q9NZI7 | ||||
| UNC4_HUMAN.H11MO.0.D |  |
HUMAN:UNCX | 11 | D | 0 | TAATYTAATTA | HOCOMOCOv10 | HT-SELEX | 45763 | Paired-related HD factors{3.1.3} | UNCX{3.1.3.27} | 33194 | 340260 | UNC4_HUMAN | A6NJT0 | ||||
| USF1_HUMAN.H11MO.0.A |  |
HUMAN:USF1 | 12 | A | 0 | vGTCACGTGvbb | HOCOMOCOv11 | ChIP-Seq | 0.998 | 0.98 | 59 | 7 | 485 | bHLH-ZIP factors{1.2.6} | USF factors{1.2.6.2} | 12593 | 7391 | USF1_HUMAN | P22415 |
| USF2_HUMAN.H11MO.0.A |  |
HUMAN:USF2 | 19 | A | 0 | vvGTCACGTGvbvvbbvbb | HOCOMOCOv10 | ChIP-Seq | 0.998 | 0.998 | 23 | 10 | 501 | bHLH-ZIP factors{1.2.6} | USF factors{1.2.6.2} | 12594 | 7392 | USF2_HUMAN | Q15853 |
| VAX1_HUMAN.H11MO.0.D |  |
HUMAN:VAX1 | 18 | D | 0 | dvTAATTARbvKWAWTWd | HOCOMOCOv10 | HT-SELEX | 50 | NK-related factors{3.1.2} | VAX{3.1.2.22} | 12660 | 11023 | VAX1_HUMAN | Q5SQQ9 | ||||
| VAX2_HUMAN.H11MO.0.D |  |
HUMAN:VAX2 | 16 | D | 0 | dndddvbTAATTARbR | HOCOMOCOv10 | HT-SELEX | 51 | NK-related factors{3.1.2} | VAX{3.1.2.22} | 12661 | 25806 | VAX2_HUMAN | Q9UIW0 | ||||
| VDR_HUMAN.H11MO.0.A | 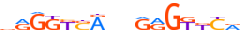 |
HUMAN:VDR | 16 | A | 0 | vRGKKSAbhRRGKKYR | HOCOMOCOv11 | ChIP-Seq | 0.943 | 0.932 | 48 | 30 | 500 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 12679 | 7421 | VDR_HUMAN | P11473 |
| VDR_HUMAN.H11MO.1.A |  |
HUMAN:VDR | 9 | A | 1 | hRRGKTCAb | HOCOMOCOv11 | ChIP-Seq | 0.877 | 0.821 | 48 | 30 | 500 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 12679 | 7421 | VDR_HUMAN | P11473 |
| VENTX_HUMAN.H11MO.0.D | 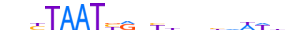 |
HUMAN:VENTX | 19 | D | 0 | bvYTAATYRbYhhhhhWWd | HOCOMOCOv10 | HT-SELEX | 9943 | NK-related factors{3.1.2} | VENTX{3.1.2.23} | 13639 | 27287 | VENTX_HUMAN | O95231 | ||||
| VEZF1_HUMAN.H11MO.0.C |  |
HUMAN:VEZF1 | 22 | C | 0 | dRRRRdRdRGGARRRRRvdRRR | HOCOMOCOv11 | ChIP-Seq | 0.909 | 5 | 500 | Factors with multiple dispersed zinc fingers{2.3.4} | MAZ-like factors{2.3.4.8} | 12949 | 7716 | VEZF1_HUMAN | Q14119 | ||
| VEZF1_HUMAN.H11MO.1.C |  |
HUMAN:VEZF1 | 12 | C | 1 | dvnRGGARRdRv | HOCOMOCOv11 | ChIP-Seq | 0.87 | 5 | 489 | Factors with multiple dispersed zinc fingers{2.3.4} | MAZ-like factors{2.3.4.8} | 12949 | 7716 | VEZF1_HUMAN | Q14119 | ||
| VSX1_HUMAN.H11MO.0.D |  |
HUMAN:VSX1 | 14 | D | 0 | TAATTARhhhhhhd | HOCOMOCOv10 | HT-SELEX | 6598 | Paired-related HD factors{3.1.3} | VSX{3.1.3.28} | 12723 | 30813 | VSX1_HUMAN | Q9NZR4 | ||||
| VSX2_HUMAN.H11MO.0.D |  |
HUMAN:VSX2 | 12 | D | 0 | hTAATTAGCYdn | HOCOMOCOv11 | ChIP-Seq | 0.996 | 3 | 504 | Paired-related HD factors{3.1.3} | VSX{3.1.3.28} | 1975 | 338917 | VSX2_HUMAN | P58304 | ||
| WT1_HUMAN.H11MO.0.C |  |
HUMAN:WT1 | 20 | C | 0 | vvSnGRRGGAGGRRvvRRvR | HOCOMOCOv11 | ChIP-Seq | 0.985 | 5 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 12796 | 7490 | WT1_HUMAN | P19544 | ||
| WT1_HUMAN.H11MO.1.B | 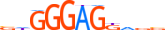 |
HUMAN:WT1 | 11 | B | 1 | vdGGGAGKddv | HOCOMOCOv11 | ChIP-Seq | 0.986 | 5 | 115 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 12796 | 7490 | WT1_HUMAN | P19544 | ||
| XBP1_HUMAN.H11MO.0.D |  |
HUMAN:XBP1 | 11 | D | 0 | RvdSASGTGKM | HOCOMOCOv11 | ChIP-Seq | 0.795 | 5 | 502 | XBP-1-related factors{1.1.5} | XBP-1{1.1.5.0.1} | 12801 | 7494 | XBP1_HUMAN | P17861 | ||
| Z324A_HUMAN.H11MO.0.C |  |
HUMAN:ZNF324 | 23 | C | 0 | bvYYvMvCYhKCCYYdbMhbbMh | HOCOMOCOv11 | ChIP-Seq | 0.986 | 3 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF324 factors{2.3.3.36} | 14096 | 25799 | Z324A_HUMAN | O75467 | ||
| Z354A_HUMAN.H11MO.0.C |  |
HUMAN:ZNF354A | 24 | C | 0 | dYYYhddYYhYTTMYWYYYRdTRW | HOCOMOCOv11 | ChIP-Seq | 0.894 | 3 | 496 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF354A-like factors{2.3.3.64} | 11628 | 6940 | Z354A_HUMAN | O60765 | ||
| ZBED1_HUMAN.H11MO.0.D |  |
HUMAN:ZBED1 | 13 | D | 0 | YATGTCGCGATAb | HOCOMOCOv10 | HT-SELEX | 4698 | BED zinc finger factors{2.3.5} | ZBED1 (ALTE, DREF, TRAMP){2.3.5.0.1} | 447 | 9189 | ZBED1_HUMAN | O96006 | ||||
| ZBT14_HUMAN.H11MO.0.C |  |
HUMAN:ZBTB14 | 9 | C | 0 | vSAGCGCGS | HOCOMOCOv11 | ChIP-Seq | 0.812 | 3 | 481 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 12860 | 7541 | ZBT14_HUMAN | O43829 | ||
| ZBT17_HUMAN.H11MO.0.A |  |
HUMAN:ZBTB17 | 19 | A | 0 | vndRvhGRRGGMdGRRvdd | HOCOMOCOv11 | ChIP-Seq | 0.959 | 0.911 | 21 | 25 | 513 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 12936 | 7709 | ZBT17_HUMAN | Q13105 |
| ZBT18_HUMAN.H11MO.0.C |  |
HUMAN:ZBTB18 | 11 | C | 0 | bSCAGhTGYKb | HOCOMOCOv11 | ChIP-Seq | 0.886 | 4 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF238-like factors{2.3.3.16} | 13030 | 10472 | ZBT18_HUMAN | Q99592 | ||
| ZBT48_HUMAN.H11MO.0.C |  |
HUMAN:ZBTB48 | 12 | C | 0 | YhAGGGAbYvdv | HOCOMOCOv11 | ChIP-Seq | 0.885 | 2 | 501 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 4930 | 3104 | ZBT48_HUMAN | P10074 | ||
| ZBT49_HUMAN.H11MO.0.D |  |
HUMAN:ZBTB49 | 11 | D | 0 | nnvnTAATYdv | HOCOMOCOv10 | HT-SELEX | 1382 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 19883 | 166793 | ZBT49_HUMAN | Q6ZSB9 | ||||
| ZBT7A_HUMAN.H11MO.0.A |  |
HUMAN:ZBTB7A | 9 | A | 0 | vRGGGKCbY | HOCOMOCOv11 | ChIP-Seq | 0.929 | 12 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | ZBTB7 factors{2.3.3.8} | 18078 | 51341 | ZBT7A_HUMAN | O95365 | ||
| ZBT7B_HUMAN.H11MO.0.D | 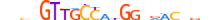 |
HUMAN:ZBTB7B | 21 | D | 0 | bbbbGTTGCCRdGGnRMCvvv | HOCOMOCOv10 | ChIP-Seq | 405 | More than 3 adjacent zinc finger factors{2.3.3} | ZBTB7 factors{2.3.3.8} | 18668 | 51043 | ZBT7B_HUMAN | O15156 | ||||
| ZBTB4_HUMAN.H11MO.0.D |  |
HUMAN:ZBTB4 | 17 | D | 0 | nCWAGATGGCWGGTGGG | HOCOMOCOv9 | Integrative | 19 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 23847 | 57659 | ZBTB4_HUMAN | Q9P1Z0 | ||||
| ZBTB4_HUMAN.H11MO.1.D | 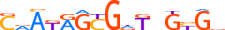 |
HUMAN:ZBTB4 | 15 | D | 1 | CRAKAGCGRTdGYGn | HOCOMOCOv9 | Integrative | 32 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 23847 | 57659 | ZBTB4_HUMAN | Q9P1Z0 | ||||
| ZBTB6_HUMAN.H11MO.0.C |  |
HUMAN:ZBTB6 | 13 | C | 0 | KRhSMTRGAGMYd | HOCOMOCOv11 | ChIP-Seq | 0.884 | 2 | 457 | More than 3 adjacent zinc finger factors{2.3.3} | ZBTB6-like factors{2.3.3.11} | 16764 | 10773 | ZBTB6_HUMAN | Q15916 | ||
| ZEB1_HUMAN.H11MO.0.A |  |
HUMAN:ZEB1 | 10 | A | 0 | bvCAGGTRdR | HOCOMOCOv11 | ChIP-Seq | 0.938 | 12 | 500 | HD-ZF factors{3.1.8} | ZEB{3.1.8.3} | 11642 | 6935 | ZEB1_HUMAN | P37275 | ||
| ZEP1_HUMAN.H11MO.0.D |  |
HUMAN:HIVEP1 | 12 | D | 0 | KGGGAAATCCCn | HOCOMOCOv9 | Integrative | 10 | Factors with multiple dispersed zinc fingers{2.3.4} | HIV EP-factors{2.3.4.5} | 4920 | 3096 | ZEP1_HUMAN | P15822 | ||||
| ZEP2_HUMAN.H11MO.0.D |  |
HUMAN:HIVEP2 | 14 | D | 0 | KSYdbGGWAASnSS | HOCOMOCOv9 | Integrative | 16 | Factors with multiple dispersed zinc fingers{2.3.4} | HIV EP-factors{2.3.4.5} | 4921 | 3097 | ZEP2_HUMAN | P31629 | ||||
| ZF64A_HUMAN.H11MO.0.D |  |
HUMAN:ZFP64 | 16 | D | 0 | vSvSCCCRGGRvbvnS | HOCOMOCOv11 | ChIP-Seq | 0.785 | 2 | 483 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 15940 | 55734 | ZF64A_HUMAN | Q9NPA5 | ||
| ZFHX3_HUMAN.H11MO.0.D |  |
HUMAN:ZFHX3 | 11 | D | 0 | RTTAATWATTW | HOCOMOCOv9 | Integrative | 7 | HD-ZF factors{3.1.8} | ZFHX{3.1.8.4} | 777 | 463 | ZFHX3_HUMAN | Q15911 | ||||
| ZFP28_HUMAN.H11MO.0.C |  |
HUMAN:ZFP28 | 20 | C | 0 | YTCYRTTTCYTCWKRWKTSA | HOCOMOCOv11 | ChIP-Seq | 0.993 | 3 | 417 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 17801 | 140612 | ZFP28_HUMAN | Q8NHY6 | ||
| ZFP42_HUMAN.H11MO.0.A |  |
HUMAN:ZFP42 | 12 | A | 0 | vRShGCCATYTY | HOCOMOCOv11 | ChIP-Seq | 0.909 | 0.8 | 3 | 2 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | YY1-like factors{2.3.3.9} | 30949 | 132625 | ZFP42_HUMAN | Q96MM3 |
| ZFP82_HUMAN.H11MO.0.C |  |
HUMAN:ZFP82 | 24 | C | 0 | YTCChdYYbWbbRWdWYTYYYTYY | HOCOMOCOv11 | ChIP-Seq | 0.93 | 3 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | ZFP30-like factors{2.3.3.63} | 28682 | 284406 | ZFP82_HUMAN | Q8N141 | ||
| ZFX_HUMAN.H11MO.0.A |  |
HUMAN:ZFX | 10 | A | 0 | vbAGGCCbvR | HOCOMOCOv11 | ChIP-Seq | 0.827 | 0.991 | 10 | 4 | 492 | More than 3 adjacent zinc finger factors{2.3.3} | ZFX/ZFY factors{2.3.3.65} | 12869 | 7543 | ZFX_HUMAN | P17010 |
| ZFX_HUMAN.H11MO.1.A | 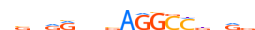 |
HUMAN:ZFX | 17 | A | 1 | dbvRRvvvAGGCMbvRv | HOCOMOCOv11 | ChIP-Seq | 0.807 | 0.957 | 10 | 4 | 495 | More than 3 adjacent zinc finger factors{2.3.3} | ZFX/ZFY factors{2.3.3.65} | 12869 | 7543 | ZFX_HUMAN | P17010 |
| ZIC1_HUMAN.H11MO.0.B |  |
HUMAN:ZIC1 | 9 | B | 0 | KGGGWGSKv | HOCOMOCOv9 | Integrative | 34 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 12872 | 7545 | ZIC1_HUMAN | Q15915 | ||||
| ZIC2_HUMAN.H11MO.0.D |  |
HUMAN:ZIC2 | 15 | D | 0 | dvhhhCCYGCdGdKn | HOCOMOCOv11 | ChIP-Seq | 0.544 | 0.977 | 4 | 4 | 410 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 12873 | 7546 | ZIC2_HUMAN | O95409 |
| ZIC3_HUMAN.H11MO.0.B |  |
HUMAN:ZIC3 | 15 | B | 0 | dvvbhCCTGCTGdGn | HOCOMOCOv11 | ChIP-Seq | 0.943 | 5 | 170 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 12874 | 7547 | ZIC3_HUMAN | O60481 | ||
| ZIC4_HUMAN.H11MO.0.D |  |
HUMAN:ZIC4 | 15 | D | 0 | dMdCMGCGGGGGKYv | HOCOMOCOv10 | HT-SELEX | 4313 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 20393 | 84107 | ZIC4_HUMAN | Q8N9L1 | ||||
| ZIM3_HUMAN.H11MO.0.C |  |
HUMAN:ZIM3 | 19 | C | 0 | bdRGvTTTCTGTTdhhnbh | HOCOMOCOv11 | ChIP-Seq | 0.989 | 3 | 471 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 16366 | 114026 | ZIM3_HUMAN | Q96PE6 | ||
| ZKSC1_HUMAN.H11MO.0.B |  |
HUMAN:ZKSCAN1 | 19 | B | 0 | dvhdbhCCTACTRWGYGYh | HOCOMOCOv11 | ChIP-Seq | 0.961 | 6 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF24-like factors{2.3.3.10} | 13101 | 7586 | ZKSC1_HUMAN | P17029 | ||
| ZKSC3_HUMAN.H11MO.0.D |  |
HUMAN:ZKSCAN3 | 11 | D | 0 | bbYRAGGYTAd | HOCOMOCOv10 | HT-SELEX | 138 | More than 3 adjacent zinc finger factors{2.3.3} | ZKSCAN3-like factors{2.3.3.26} | 13853 | 80317 | ZKSC3_HUMAN | Q9BRR0 | ||||
| ZN121_HUMAN.H11MO.0.C |  |
HUMAN:ZNF121 | 20 | C | 0 | CKGGGCAAYAbAGYRAGACY | HOCOMOCOv11 | ChIP-Seq | 0.92 | 2 | 486 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 12904 | 7675 | ZN121_HUMAN | P58317 | ||
| ZN134_HUMAN.H11MO.0.C |  |
HUMAN:ZNF134 | 22 | C | 0 | KhvdbvhYYMAKCAGKKGMdGS | HOCOMOCOv11 | ChIP-Seq | 0.923 | 4 | 500 | Factors with multiple dispersed zinc fingers{2.3.4} | ZNF134-like factors{2.3.4.24} | 12918 | 7693 | ZN134_HUMAN | P52741 | ||
| ZN134_HUMAN.H11MO.1.C |  |
HUMAN:ZNF134 | 17 | C | 1 | vhYYvAKCAGKKGMdGS | HOCOMOCOv11 | ChIP-Seq | 0.93 | 4 | 500 | Factors with multiple dispersed zinc fingers{2.3.4} | ZNF134-like factors{2.3.4.24} | 12918 | 7693 | ZN134_HUMAN | P52741 | ||
| ZN136_HUMAN.H11MO.0.C |  |
HUMAN:ZNF136 | 24 | C | 0 | TRCTGGRTAYARWATTCTKGGYTG | HOCOMOCOv11 | ChIP-Seq | 0.907 | 3 | 365 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF763-like factors{2.3.3.33} | 12920 | 7695 | ZN136_HUMAN | P52737 | ||
| ZN140_HUMAN.H11MO.0.C |  |
HUMAN:ZNF140 | 24 | C | 0 | hvddvnbYWKCAATTCYRCTCMhh | HOCOMOCOv11 | ChIP-Seq | 0.984 | 4 | 450 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF302-like factors{2.3.3.44} | 12925 | 7699 | ZN140_HUMAN | P52738 | ||
| ZN143_HUMAN.H11MO.0.A |  |
HUMAN:ZNF143 | 22 | A | 0 | dGSMYbMTGGGAddYGTAGTYY | HOCOMOCOv11 | ChIP-Seq | 0.986 | 0.983 | 28 | 9 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF76-like factors{2.3.3.28} | 12928 | 7702 | ZN143_HUMAN | P52747 |
| ZN148_HUMAN.H11MO.0.D |  |
HUMAN:ZNF148 | 15 | D | 0 | dSvKbGGGGGhGGGR | HOCOMOCOv9 | Integrative | 34 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF148-like factors{2.3.3.13} | 12933 | 7707 | ZN148_HUMAN | Q9UQR1 | ||||
| ZN214_HUMAN.H11MO.0.C |  |
HUMAN:ZNF214 | 22 | C | 0 | hhvWTbYKRdKSMhbTYSATKW | HOCOMOCOv11 | ChIP-Seq | 0.926 | 2 | 264 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF214-like factors{2.3.3.56} | 13006 | 7761 | ZN214_HUMAN | Q9UL59 | ||
| ZN219_HUMAN.H11MO.0.D |  |
HUMAN:ZNF219 | 12 | D | 0 | bdGGGGGGYSGW | HOCOMOCOv9 | Integrative | 26 | Factors with multiple dispersed zinc fingers{2.3.4} | ZNF219-like factors{2.3.4.2} | 13011 | 51222 | ZN219_HUMAN | Q9P2Y4 | ||||
| ZN232_HUMAN.H11MO.0.D |  |
HUMAN:ZNF232 | 19 | D | 0 | nGTTAAAYGTAGATTAARh | HOCOMOCOv10 | HT-SELEX | 50 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF24-like factors{2.3.3.10} | 13026 | 7775 | ZN232_HUMAN | Q9UNY5 | ||||
| ZN250_HUMAN.H11MO.0.C |  |
HUMAN:ZNF250 | 20 | C | 0 | TACCATRCTGTTTTGRTTAC | HOCOMOCOv11 | ChIP-Seq | 0.904 | 4 | 360 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 13044 | 58500 | ZN250_HUMAN | P15622 | ||
| ZN257_HUMAN.H11MO.0.C |  |
HUMAN:ZNF257 | 12 | C | 0 | KhYCTTGCCWCh | HOCOMOCOv11 | ChIP-Seq | 0.867 | 4 | 432 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 13498 | 113835 | ZN257_HUMAN | Q9Y2Q1 | ||
| ZN260_HUMAN.H11MO.0.C |  |
HUMAN:ZNF260 | 24 | C | 0 | TTTdYATRGCTGYATASTATTCCA | HOCOMOCOv11 | ChIP-Seq | 0.921 | 4 | 443 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF146-like factors{2.3.3.55} | 13499 | 339324 | ZN260_HUMAN | Q3ZCT1 | ||
| ZN263_HUMAN.H11MO.0.A |  |
HUMAN:ZNF263 | 20 | A | 0 | RGGAGGASSvvSvRRvvvnv | HOCOMOCOv11 | ChIP-Seq | 0.968 | 9 | 501 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 13056 | 10127 | ZN263_HUMAN | O14978 | ||
| ZN263_HUMAN.H11MO.1.A |  |
HUMAN:ZNF263 | 11 | A | 1 | vdGGGAGGASK | HOCOMOCOv11 | ChIP-Seq | 0.985 | 9 | 505 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 13056 | 10127 | ZN263_HUMAN | O14978 | ||
| ZN264_HUMAN.H11MO.0.C |  |
HUMAN:ZNF264 | 24 | C | 0 | bhYbvhMWRGRCWCKRATYbhMYb | HOCOMOCOv11 | ChIP-Seq | 0.812 | 2 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF460-like factors{2.3.3.54} | 13057 | 9422 | ZN264_HUMAN | O43296 | ||
| ZN274_HUMAN.H11MO.0.A |  |
HUMAN:ZNF274 | 20 | A | 0 | CAYMMWGGWRWGRARYSbWA | HOCOMOCOv11 | ChIP-Seq | 0.975 | 26 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 13068 | 10782 | ZN274_HUMAN | Q96GC6 | ||
| ZN281_HUMAN.H11MO.0.A |  |
HUMAN:ZNF281 | 15 | A | 0 | vRvdGGGGGWdGGGv | HOCOMOCOv11 | ChIP-Seq | 0.968 | 0.972 | 7 | 3 | 482 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF148-like factors{2.3.3.13} | 13075 | 23528 | ZN281_HUMAN | Q9Y2X9 |
| ZN282_HUMAN.H11MO.0.D |  |
HUMAN:ZNF282 | 14 | D | 0 | nRKRTnAKGSGARW | HOCOMOCOv10 | HT-SELEX | 36 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF282-like factors{2.3.3.38} | 13076 | 8427 | ZN282_HUMAN | Q9UDV7 | ||||
| ZN317_HUMAN.H11MO.0.C |  |
HUMAN:ZNF317 | 20 | C | 0 | RRhWRACWRMWGAYWWYYbR | HOCOMOCOv11 | ChIP-Seq | 0.905 | 2 | 455 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 13507 | 57693 | ZN317_HUMAN | Q96PQ6 | ||
| ZN320_HUMAN.H11MO.0.C |  |
HUMAN:ZNF320 | 20 | C | 0 | KKKGGdYMhRKGGGCMddbK | HOCOMOCOv11 | ChIP-Seq | 0.981 | 3 | 461 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 13842 | 162967 | ZN320_HUMAN | A2RRD8 | ||
| ZN322_HUMAN.H11MO.0.B |  |
HUMAN:ZNF322 | 20 | B | 0 | RdKMCYRbYMCdSdGCYKSv | HOCOMOCOv11 | ChIP-Seq | 0.884 | 0.713 | 4 | 2 | 465 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF322-like factors{2.3.3.52} | 23640 | 79692 | ZN322_HUMAN | Q6U7Q0 |
| ZN329_HUMAN.H11MO.0.C |  |
HUMAN:ZNF329 | 20 | C | 0 | CTKGATCMARCYdYRCCTGA | HOCOMOCOv11 | ChIP-Seq | 0.828 | 4 | 361 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 14209 | 79673 | ZN329_HUMAN | Q86UD4 | ||
| ZN331_HUMAN.H11MO.0.C |  |
HUMAN:ZNF331 | 20 | C | 0 | GRYWGGGCTCAGCWGGRhRG | HOCOMOCOv11 | ChIP-Seq | 0.989 | 3 | 458 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 15489 | 55422 | ZN331_HUMAN | Q9NQX6 | ||
| ZN333_HUMAN.H11MO.0.D |  |
HUMAN:ZNF333 | 9 | D | 0 | bKATAATKh | HOCOMOCOv9 | Integrative | 16 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF177-like factors{2.3.3.40} | 15624 | 84449 | ZN333_HUMAN | Q96JL9 | ||||
| ZN335_HUMAN.H11MO.0.A |  |
HUMAN:ZNF335 | 22 | A | 0 | RKSTGYCYnnnbhdvTGCCTSh | HOCOMOCOv11 | ChIP-Seq | 0.96 | 0.968 | 2 | 4 | 103 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 15807 | 63925 | ZN335_HUMAN | Q9H4Z2 |
| ZN335_HUMAN.H11MO.1.A |  |
HUMAN:ZNF335 | 7 | A | 1 | YGCCTGW | HOCOMOCOv11 | ChIP-Seq | 0.892 | 0.995 | 2 | 4 | 123 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 15807 | 63925 | ZN335_HUMAN | Q9H4Z2 |
| ZN341_HUMAN.H11MO.0.C |  |
HUMAN:ZNF341 | 22 | C | 0 | dvvRvRvdRvRRddGGAASRGM | HOCOMOCOv11 | ChIP-Seq | 0.845 | 2 | 499 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 15992 | 84905 | ZN341_HUMAN | Q9BYN7 | ||
| ZN341_HUMAN.H11MO.1.C |  |
HUMAN:ZNF341 | 9 | C | 1 | GGAASAGCh | HOCOMOCOv11 | ChIP-Seq | 0.813 | 2 | 500 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 15992 | 84905 | ZN341_HUMAN | Q9BYN7 | ||
| ZN350_HUMAN.H11MO.0.C |  |
HUMAN:ZNF350 | 18 | C | 0 | vdbbYYYTTYhWdhYYhd | HOCOMOCOv11 | ChIP-Seq | 0.978 | 3 | 501 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF350-like factors{2.3.3.30} | 16656 | 59348 | ZN350_HUMAN | Q9GZX5 | ||
| ZN350_HUMAN.H11MO.1.D |  |
HUMAN:ZNF350 | 24 | D | 1 | nSGGGvSRMAGnnvKTTGKbKSCn | HOCOMOCOv9 | Integrative | 0.671 | 3 | 15 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF350-like factors{2.3.3.30} | 16656 | 59348 | ZN350_HUMAN | Q9GZX5 | ||
| ZN382_HUMAN.H11MO.0.C |  |
HUMAN:ZNF382 | 22 | C | 0 | RYdRKRYCKGTWSTRvTKYCYC | HOCOMOCOv11 | ChIP-Seq | 0.931 | 2 | 478 | Factors with multiple dispersed zinc fingers{2.3.4} | ZNF37A-like factors{2.3.4.25} | 17409 | 84911 | ZN382_HUMAN | Q96SR6 | ||
| ZN384_HUMAN.H11MO.0.C |  |
HUMAN:ZNF384 | 12 | C | 0 | SGAAAAAvbnbv | HOCOMOCOv9 | Integrative | 39 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF362-like factors{2.3.3.37} | 11955 | 171017 | ZN384_HUMAN | Q8TF68 | ||||
| ZN394_HUMAN.H11MO.0.C |  |
HUMAN:ZNF394 | 20 | C | 0 | nRARdRSMMdnvWhWRdRRd | HOCOMOCOv11 | ChIP-Seq | 0.802 | 3 | 375 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 18832 | 84124 | ZN394_HUMAN | Q53GI3 | ||
| ZN394_HUMAN.H11MO.1.D |  |
HUMAN:ZNF394 | 9 | D | 1 | dAMWGGARd | HOCOMOCOv11 | ChIP-Seq | 0.661 | 3 | 382 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 18832 | 84124 | ZN394_HUMAN | Q53GI3 | ||
| ZN410_HUMAN.H11MO.0.D |  |
HUMAN:ZNF410 | 13 | D | 0 | TATTATGGGATGb | HOCOMOCOv10 | HT-SELEX | 1227 | More than 3 adjacent zinc finger factors{2.3.3} | MTF1-like factors{2.3.3.24} | 20144 | 57862 | ZN410_HUMAN | Q86VK4 | ||||
| ZN418_HUMAN.H11MO.0.C |  |
HUMAN:ZNF418 | 17 | C | 0 | TGYTTYYRKYYdMYbnb | HOCOMOCOv11 | ChIP-Seq | 0.906 | 4 | 500 | Factors with multiple dispersed zinc fingers{2.3.4} | ZNF417-like factors{2.3.4.1} | 20647 | 147686 | ZN418_HUMAN | Q8TF45 | ||
| ZN418_HUMAN.H11MO.1.D |  |
HUMAN:ZNF418 | 12 | D | 1 | dvhTGMTTYYRK | HOCOMOCOv11 | ChIP-Seq | 0.791 | 4 | 500 | Factors with multiple dispersed zinc fingers{2.3.4} | ZNF417-like factors{2.3.4.1} | 20647 | 147686 | ZN418_HUMAN | Q8TF45 | ||
| ZN423_HUMAN.H11MO.0.D |  |
HUMAN:ZNF423 | 15 | D | 0 | GGCACCCAAGGGTGC | HOCOMOCOv9 | Integrative | 22 | Factors with multiple dispersed zinc fingers{2.3.4} | ZNF423-like factors{2.3.4.12} | 16762 | 23090 | ZN423_HUMAN | Q2M1K9 | ||||
| ZN436_HUMAN.H11MO.0.C |  |
HUMAN:ZNF436 | 24 | C | 0 | KCMdGGARGRCTTCCTGGAGGAGG | HOCOMOCOv11 | ChIP-Seq | 0.984 | 4 | 485 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF180-like factors{2.3.3.58} | 20814 | 80818 | ZN436_HUMAN | Q9C0F3 | ||
| ZN449_HUMAN.H11MO.0.C |  |
HUMAN:ZNF449 | 11 | C | 0 | dGGYTGGGCWb | HOCOMOCOv11 | ChIP-Seq | 0.938 | 2 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 21039 | 203523 | ZN449_HUMAN | Q6P9G9 | ||
| ZN467_HUMAN.H11MO.0.C |  |
HUMAN:ZNF467 | 22 | C | 0 | RGGGAGGRRvvRRRRRRRRRRv | HOCOMOCOv11 | ChIP-Seq | 0.971 | 3 | 499 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 23154 | 168544 | ZN467_HUMAN | Q7Z7K2 | ||
| ZN490_HUMAN.H11MO.0.C |  |
HUMAN:ZNF490 | 24 | C | 0 | TRdGTYTCTTGWAGRCAGCAKAYM | HOCOMOCOv11 | ChIP-Seq | 0.989 | 4 | 430 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF763-like factors{2.3.3.33} | 23705 | 57474 | ZN490_HUMAN | Q9ULM2 | ||
| ZN502_HUMAN.H11MO.0.C |  |
HUMAN:ZNF502 | 20 | C | 0 | GAATGGAATSGAATGGAATS | HOCOMOCOv11 | ChIP-Seq | 0.876 | 3 | 156 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 23718 | 91392 | ZN502_HUMAN | Q8TBZ5 | ||
| ZN524_HUMAN.H11MO.0.D |  |
HUMAN:ZNF524 | 18 | D | 0 | bSKdnbdMGGGTKCGARb | HOCOMOCOv10 | HT-SELEX | 509 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF524-like factors{2.3.3.15} | 28322 | 147807 | ZN524_HUMAN | Q96C55 | ||||
| ZN528_HUMAN.H11MO.0.C |  |
HUMAN:ZNF528 | 20 | C | 0 | nnvRRGGAAGYCATTYCTbd | HOCOMOCOv11 | ChIP-Seq | 0.82 | 2 | 371 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 29384 | 84436 | ZN528_HUMAN | Q3MIS6 | ||
| ZN547_HUMAN.H11MO.0.C |  |
HUMAN:ZNF547 | 20 | C | 0 | WSCWMAYKChRCWRRCRbdS | HOCOMOCOv11 | ChIP-Seq | 0.901 | 4 | 405 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 26432 | 284306 | ZN547_HUMAN | Q8IVP9 | ||
| ZN549_HUMAN.H11MO.0.C |  |
HUMAN:ZNF549 | 20 | C | 0 | WdYYAKSRRYhGGRCAGCMb | HOCOMOCOv11 | ChIP-Seq | 0.948 | 3 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 26632 | 256051 | ZN549_HUMAN | Q6P9A3 | ||
| ZN554_HUMAN.H11MO.0.C | 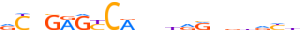 |
HUMAN:ZNF554 | 20 | C | 0 | RCdGRGYCRnbbRRdddvYh | HOCOMOCOv11 | ChIP-Seq | 0.853 | 2 | 501 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 26629 | 115196 | ZN554_HUMAN | Q86TJ5 | ||
| ZN554_HUMAN.H11MO.1.D |  |
HUMAN:ZNF554 | 9 | D | 1 | RCWGAGYCM | HOCOMOCOv11 | ChIP-Seq | 0.774 | 2 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 26629 | 115196 | ZN554_HUMAN | Q86TJ5 | ||
| ZN563_HUMAN.H11MO.0.C |  |
HUMAN:ZNF563 | 21 | C | 0 | dKdnKCMbndShRKCMvChSh | HOCOMOCOv11 | ChIP-Seq | 0.886 | 4 | 499 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF763-like factors{2.3.3.33} | 30498 | 147837 | ZN563_HUMAN | Q8TA94 | ||
| ZN563_HUMAN.H11MO.1.C |  |
HUMAN:ZNF563 | 12 | C | 1 | ShRKCAGCdShR | HOCOMOCOv11 | ChIP-Seq | 0.863 | 4 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF763-like factors{2.3.3.33} | 30498 | 147837 | ZN563_HUMAN | Q8TA94 | ||
| ZN582_HUMAN.H11MO.0.C |  |
HUMAN:ZNF582 | 24 | C | 0 | vKMWTGSWnTbGRMTGSAAdYAWM | HOCOMOCOv11 | ChIP-Seq | 0.852 | 3 | 261 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF620-like factors{2.3.3.35} | 26421 | 147948 | ZN582_HUMAN | Q96NG8 | ||
| ZN586_HUMAN.H11MO.0.C |  |
HUMAN:ZNF586 | 20 | C | 0 | AGGCCTAGGAGGAAAAAATG | HOCOMOCOv11 | ChIP-Seq | 1.0 | 4 | 472 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 25949 | 54807 | ZN586_HUMAN | Q9NXT0 | ||
| ZN589_HUMAN.H11MO.0.D |  |
HUMAN:ZNF589 | 16 | D | 0 | nMMRSKGKTWYWRYSb | HOCOMOCOv9 | Integrative | 15 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 16747 | 51385 | ZN589_HUMAN | Q86UQ0 | ||||
| ZN652_HUMAN.H11MO.0.D |  |
HUMAN:ZNF652 | 11 | D | 0 | nRdKGGTTAAh | HOCOMOCOv10 | HT-SELEX | 236 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF652-like factors{2.3.3.29} | 29147 | 22834 | ZN652_HUMAN | Q9Y2D9 | ||||
| ZN667_HUMAN.H11MO.0.C |  |
HUMAN:ZNF667 | 19 | C | 0 | vdKbChKbMRRWKYYShbh | HOCOMOCOv11 | ChIP-Seq | 0.809 | 3 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 28854 | 63934 | ZN667_HUMAN | Q5HYK9 | ||
| ZN680_HUMAN.H11MO.0.C |  |
HUMAN:ZNF680 | 20 | C | 0 | YRYRhCCARGAAGAATRAGG | HOCOMOCOv11 | ChIP-Seq | 0.999 | 3 | 309 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 26897 | 340252 | ZN680_HUMAN | Q8NEM1 | ||
| ZN708_HUMAN.H11MO.0.C |  |
HUMAN:ZNF708 | 20 | C | 0 | vvvhhddMRRnAGGYACAGC | HOCOMOCOv11 | ChIP-Seq | 0.838 | 3 | 359 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 12945 | 7562 | ZN708_HUMAN | P17019 | ||
| ZN708_HUMAN.H11MO.1.D |  |
HUMAN:ZNF708 | 9 | D | 1 | AGGYACAGM | HOCOMOCOv11 | ChIP-Seq | 0.795 | 3 | 439 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 12945 | 7562 | ZN708_HUMAN | P17019 | ||
| ZN713_HUMAN.H11MO.0.D |  |
HUMAN:ZNF713 | 19 | D | 0 | ddRRMMRAAnGvSAnKMWd | HOCOMOCOv10 | HT-SELEX | 222 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 22043 | ZN713_HUMAN | Q8N859 | |||||
| ZN740_HUMAN.H11MO.0.D |  |
HUMAN:ZNF740 | 14 | D | 0 | ddKGGGGGGGhddd | HOCOMOCOv10 | HT-SELEX | 2301 | Other factors with up to three adjacent zinc fingers{2.3.2} | Other three adjacent zinc finger factors{2.3.2.4} | 27465 | 283337 | ZN740_HUMAN | Q8NDX6 | ||||
| ZN768_HUMAN.H11MO.0.C |  |
HUMAN:ZNF768 | 20 | C | 0 | vRRGMWCAGAGAGGKbRRGb | HOCOMOCOv11 | ChIP-Seq | 0.881 | 4 | 424 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 26273 | 79724 | ZN768_HUMAN | Q9H5H4 | ||
| ZN770_HUMAN.H11MO.0.C |  |
HUMAN:ZNF770 | 22 | C | 0 | dGGAGGCYRRRRbdRRRGRvbb | HOCOMOCOv11 | ChIP-Seq | 0.969 | 3 | 491 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 26061 | 54989 | ZN770_HUMAN | Q6IQ21 | ||
| ZN770_HUMAN.H11MO.1.C |  |
HUMAN:ZNF770 | 11 | C | 1 | dGGAGGCYGvR | HOCOMOCOv11 | ChIP-Seq | 0.962 | 3 | 491 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 26061 | 54989 | ZN770_HUMAN | Q6IQ21 | ||
| ZN784_HUMAN.H11MO.0.D |  |
HUMAN:ZNF784 | 11 | D | 0 | dvGGTAGGTvn | HOCOMOCOv10 | HT-SELEX | 256 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 33111 | 147808 | ZN784_HUMAN | Q8NCA9 | ||||
| ZN816_HUMAN.H11MO.0.C |  |
HUMAN:ZNF816 | 21 | C | 0 | vRvdvRdGGGGACMKGhhdRR | HOCOMOCOv11 | ChIP-Seq | 0.95 | 4 | 430 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF816A-like factors{2.3.3.73} | 26995 | 125893 | ZN816_HUMAN | Q0VGE8 | ||
| ZN816_HUMAN.H11MO.1.C |  |
HUMAN:ZNF816 | 14 | C | 1 | vddRGGGAMWdRhd | HOCOMOCOv11 | ChIP-Seq | 0.888 | 4 | 464 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF816A-like factors{2.3.3.73} | 26995 | 125893 | ZN816_HUMAN | Q0VGE8 | ||
| ZNF18_HUMAN.H11MO.0.C |  |
HUMAN:ZNF18 | 12 | C | 0 | RGTGTGARYhbd | HOCOMOCOv11 | ChIP-Seq | 0.952 | 4 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 12969 | 7566 | ZNF18_HUMAN | P17022 | ||
| ZNF41_HUMAN.H11MO.0.C |  |
HUMAN:ZNF41 | 24 | C | 0 | vRMRRGRRARhvvRvSACMAWSWR | HOCOMOCOv11 | ChIP-Seq | 0.894 | 4 | 349 | More than 3 adjacent zinc finger factors{2.3.3} | ZFN81-like factors{2.3.3.68} | 13107 | 7592 | ZNF41_HUMAN | P51814 | ||
| ZNF41_HUMAN.H11MO.1.C |  |
HUMAN:ZNF41 | 12 | C | 1 | MAYARGGRARhv | HOCOMOCOv11 | ChIP-Seq | 0.854 | 4 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | ZFN81-like factors{2.3.3.68} | 13107 | 7592 | ZNF41_HUMAN | P51814 | ||
| ZNF76_HUMAN.H11MO.0.C |  |
HUMAN:ZNF76 | 22 | C | 0 | vddGSMYbSTGGGAvdKSKRRK | HOCOMOCOv11 | ChIP-Seq | 0.985 | 4 | 498 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF76-like factors{2.3.3.28} | 13149 | 7629 | ZNF76_HUMAN | P36508 | ||
| ZNF85_HUMAN.H11MO.0.C |  |
HUMAN:ZNF85 | 20 | C | 0 | RdWRRMRWvKGMWGbAATCW | HOCOMOCOv11 | ChIP-Seq | 0.899 | 4 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 13160 | 7639 | ZNF85_HUMAN | Q03923 | ||
| ZNF85_HUMAN.H11MO.1.C |  |
HUMAN:ZNF85 | 13 | C | 1 | KGMWGbAATCYhh | HOCOMOCOv11 | ChIP-Seq | 0.896 | 4 | 499 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 13160 | 7639 | ZNF85_HUMAN | Q03923 | ||
| ZNF8_HUMAN.H11MO.0.C |  |
HUMAN:ZNF8 | 22 | C | 0 | TGTGGTAYATCCATWYARTGGA | HOCOMOCOv11 | ChIP-Seq | 0.982 | 3 | 417 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 13154 | 7554 | ZNF8_HUMAN | P17098 | ||
| ZSC16_HUMAN.H11MO.0.D |  |
HUMAN:ZSCAN16 | 20 | D | 0 | nvbGTGYTCWGTTAACAhhn | HOCOMOCOv10 | HT-SELEX | 23 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 20813 | 80345 | ZSC16_HUMAN | Q9H4T2 | ||||
| ZSC22_HUMAN.H11MO.0.C |  |
HUMAN:ZSCAN22 | 16 | C | 0 | dRbShGAdGGMGGMGG | HOCOMOCOv11 | ChIP-Seq | 0.932 | 2 | 476 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 4929 | 342945 | ZSC22_HUMAN | P10073 | ||
| ZSC31_HUMAN.H11MO.0.C |  |
HUMAN:ZSCAN31 | 18 | C | 0 | dKSMGCRGGGCMdKdRdv | HOCOMOCOv11 | ChIP-Seq | 0.802 | 4 | 407 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF24-like factors{2.3.3.10} | 14097 | 64288 | ZSC31_HUMAN | Q96LW9 | ||
| ZSCA4_HUMAN.H11MO.0.D |  |
HUMAN:ZSCAN4 | 12 | D | 0 | AGKGTGTGCAhh | HOCOMOCOv10 | HT-SELEX | 1158 | More than 3 adjacent zinc finger factors{2.3.3} | ZSCAN5-like zinc finger factors{2.3.3.5} | 23709 | 201516 | ZSCA4_HUMAN | Q8NAM6 |