Model info
| Transcription factor | MAZ (GeneCards) | ||||||||
| Model | MAZ_HUMAN.H11MO.1.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 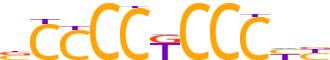 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 11 | ||||||||
| Quality | A | ||||||||
| Motif rank | 1 | ||||||||
| Consensus | RRGGGMGGGGv | ||||||||
| Best auROC (human) | 0.926 | ||||||||
| Best auROC (mouse) | 0.955 | ||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | 5 | ||||||||
| Aligned words | 486 | ||||||||
| TF family | Factors with multiple dispersed zinc fingers {2.3.4} | ||||||||
| TF subfamily | MAZ-like factors {2.3.4.8} | ||||||||
| HGNC | HGNC:6914 | ||||||||
| EntrezGene | GeneID:4150 (SSTAR profile) | ||||||||
| UniProt ID | MAZ_HUMAN | ||||||||
| UniProt AC | P56270 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | MAZ expression | ||||||||
| ReMap ChIP-seq dataset list | MAZ datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 157.0 | 67.0 | 249.0 | 13.0 |
| 02 | 237.0 | 22.0 | 179.0 | 48.0 |
| 03 | 32.0 | 3.0 | 448.0 | 3.0 |
| 04 | 0.0 | 0.0 | 481.0 | 5.0 |
| 05 | 1.0 | 3.0 | 481.0 | 1.0 |
| 06 | 315.0 | 147.0 | 0.0 | 24.0 |
| 07 | 14.0 | 1.0 | 470.0 | 1.0 |
| 08 | 2.0 | 0.0 | 481.0 | 3.0 |
| 09 | 60.0 | 8.0 | 407.0 | 11.0 |
| 10 | 60.0 | 3.0 | 421.0 | 2.0 |
| 11 | 56.0 | 166.0 | 228.0 | 36.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.253 | -0.585 | 0.711 | -2.135 |
| 02 | 0.662 | -1.654 | 0.383 | -0.91 |
| 03 | -1.3 | -3.298 | 1.296 | -3.298 |
| 04 | -4.377 | -4.377 | 1.367 | -2.934 |
| 05 | -3.878 | -3.298 | 1.367 | -3.878 |
| 06 | 0.945 | 0.188 | -4.377 | -1.572 |
| 07 | -2.069 | -3.878 | 1.343 | -3.878 |
| 08 | -3.547 | -4.377 | 1.367 | -3.298 |
| 09 | -0.693 | -2.556 | 1.2 | -2.283 |
| 10 | -0.693 | -3.298 | 1.234 | -3.547 |
| 11 | -0.76 | 0.309 | 0.624 | -1.187 |