Model info
| Transcription factor | MYOD1 (GeneCards) | ||||||||
| Model | MYOD1_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 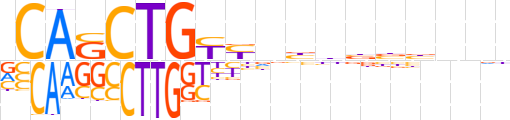 | ||||||||
| LOGO (reverse complement) | 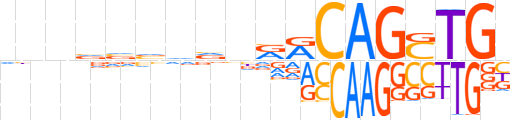 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vCASCTGYYnbbbbbnnb | ||||||||
| Best auROC (human) | 0.995 | ||||||||
| Best auROC (mouse) | 0.997 | ||||||||
| Peak sets in benchmark (human) | 9 | ||||||||
| Peak sets in benchmark (mouse) | 63 | ||||||||
| Aligned words | 437 | ||||||||
| TF family | MyoD / ASC-related factors {1.2.2} | ||||||||
| TF subfamily | Myogenic transcription factors {1.2.2.1} | ||||||||
| HGNC | HGNC:7611 | ||||||||
| EntrezGene | GeneID:4654 (SSTAR profile) | ||||||||
| UniProt ID | MYOD1_HUMAN | ||||||||
| UniProt AC | P15172 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | MYOD1 expression | ||||||||
| ReMap ChIP-seq dataset list | MYOD1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 1.0 | 122.0 | 0.0 | 0.0 | 1.0 | 112.0 | 0.0 | 1.0 | 0.0 | 161.0 | 1.0 | 1.0 | 1.0 | 36.0 | 0.0 | 0.0 |
| 02 | 3.0 | 0.0 | 0.0 | 0.0 | 415.0 | 2.0 | 3.0 | 11.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 03 | 0.0 | 137.0 | 268.0 | 14.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 4.0 | 1.0 | 0.0 | 0.0 | 3.0 | 8.0 | 0.0 |
| 04 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 144.0 | 0.0 | 2.0 | 2.0 | 269.0 | 3.0 | 3.0 | 0.0 | 14.0 | 0.0 | 0.0 |
| 05 | 0.0 | 0.0 | 0.0 | 2.0 | 4.0 | 2.0 | 0.0 | 421.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 5.0 |
| 06 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 428.0 | 3.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 166.0 | 17.0 | 246.0 | 0.0 | 0.0 | 2.0 | 2.0 |
| 08 | 1.0 | 1.0 | 1.0 | 1.0 | 11.0 | 49.0 | 1.0 | 105.0 | 0.0 | 8.0 | 3.0 | 8.0 | 3.0 | 134.0 | 29.0 | 82.0 |
| 09 | 3.0 | 5.0 | 5.0 | 2.0 | 76.0 | 44.0 | 29.0 | 43.0 | 8.0 | 17.0 | 6.0 | 3.0 | 16.0 | 61.0 | 98.0 | 21.0 |
| 10 | 15.0 | 44.0 | 25.0 | 19.0 | 17.0 | 61.0 | 6.0 | 43.0 | 6.0 | 90.0 | 10.0 | 32.0 | 12.0 | 30.0 | 18.0 | 9.0 |
| 11 | 7.0 | 13.0 | 19.0 | 11.0 | 35.0 | 58.0 | 9.0 | 123.0 | 9.0 | 21.0 | 16.0 | 13.0 | 10.0 | 25.0 | 45.0 | 23.0 |
| 12 | 6.0 | 4.0 | 45.0 | 6.0 | 32.0 | 28.0 | 15.0 | 42.0 | 15.0 | 31.0 | 33.0 | 10.0 | 6.0 | 22.0 | 126.0 | 16.0 |
| 13 | 2.0 | 17.0 | 34.0 | 6.0 | 19.0 | 37.0 | 3.0 | 26.0 | 5.0 | 105.0 | 41.0 | 68.0 | 3.0 | 23.0 | 45.0 | 3.0 |
| 14 | 2.0 | 6.0 | 18.0 | 3.0 | 37.0 | 76.0 | 6.0 | 63.0 | 8.0 | 57.0 | 41.0 | 17.0 | 2.0 | 72.0 | 20.0 | 9.0 |
| 15 | 11.0 | 12.0 | 13.0 | 13.0 | 74.0 | 53.0 | 13.0 | 71.0 | 21.0 | 26.0 | 28.0 | 10.0 | 4.0 | 34.0 | 28.0 | 26.0 |
| 16 | 35.0 | 22.0 | 28.0 | 25.0 | 49.0 | 23.0 | 15.0 | 38.0 | 21.0 | 28.0 | 17.0 | 16.0 | 21.0 | 18.0 | 61.0 | 20.0 |
| 17 | 12.0 | 16.0 | 30.0 | 68.0 | 12.0 | 25.0 | 5.0 | 49.0 | 13.0 | 18.0 | 8.0 | 82.0 | 3.0 | 33.0 | 18.0 | 45.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -2.999 | 1.486 | -4.289 | -4.289 | -2.999 | 1.401 | -4.289 | -2.999 | -4.289 | 1.763 | -2.999 | -2.999 | -2.999 | 0.273 | -4.289 | -4.289 |
| 02 | -2.103 | -4.289 | -4.289 | -4.289 | 2.708 | -2.454 | -2.103 | -0.889 | -2.999 | -4.289 | -4.289 | -4.289 | -4.289 | -4.289 | -2.454 | -4.289 |
| 03 | -4.289 | 1.602 | 2.271 | -0.655 | -4.289 | -2.454 | -4.289 | -4.289 | -4.289 | -1.844 | -2.999 | -4.289 | -4.289 | -2.103 | -1.195 | -4.289 |
| 04 | -4.289 | -4.289 | -4.289 | -4.289 | -4.289 | 1.651 | -4.289 | -2.454 | -2.454 | 2.275 | -2.103 | -2.103 | -4.289 | -0.655 | -4.289 | -4.289 |
| 05 | -4.289 | -4.289 | -4.289 | -2.454 | -1.844 | -2.454 | -4.289 | 2.722 | -4.289 | -4.289 | -4.289 | -2.103 | -4.289 | -4.289 | -4.289 | -1.638 |
| 06 | -4.289 | -4.289 | -1.844 | -4.289 | -4.289 | -4.289 | -2.999 | -2.999 | -4.289 | -4.289 | -4.289 | -4.289 | -4.289 | -4.289 | 2.739 | -2.103 |
| 07 | -4.289 | -4.289 | -4.289 | -4.289 | -4.289 | -4.289 | -4.289 | -4.289 | -1.844 | 1.793 | -0.466 | 2.186 | -4.289 | -4.289 | -2.454 | -2.454 |
| 08 | -2.999 | -2.999 | -2.999 | -2.999 | -0.889 | 0.578 | -2.999 | 1.336 | -4.289 | -1.195 | -2.103 | -1.195 | -2.103 | 1.58 | 0.059 | 1.09 |
| 09 | -2.103 | -1.638 | -1.638 | -2.454 | 1.015 | 0.472 | 0.059 | 0.449 | -1.195 | -0.466 | -1.468 | -2.103 | -0.525 | 0.796 | 1.268 | -0.259 |
| 10 | -0.588 | 0.472 | -0.087 | -0.357 | -0.466 | 0.796 | -1.468 | 0.449 | -1.468 | 1.183 | -0.981 | 0.156 | -0.805 | 0.093 | -0.41 | -1.083 |
| 11 | -1.322 | -0.727 | -0.357 | -0.889 | 0.245 | 0.746 | -1.083 | 1.494 | -1.083 | -0.259 | -0.525 | -0.727 | -0.981 | -0.087 | 0.494 | -0.169 |
| 12 | -1.468 | -1.844 | 0.494 | -1.468 | 0.156 | 0.025 | -0.588 | 0.426 | -0.588 | 0.125 | 0.187 | -0.981 | -1.468 | -0.213 | 1.518 | -0.525 |
| 13 | -2.454 | -0.466 | 0.216 | -1.468 | -0.357 | 0.3 | -2.103 | -0.049 | -1.638 | 1.336 | 0.402 | 0.904 | -2.103 | -0.169 | 0.494 | -2.103 |
| 14 | -2.454 | -1.468 | -0.41 | -2.103 | 0.3 | 1.015 | -1.468 | 0.828 | -1.195 | 0.729 | 0.402 | -0.466 | -2.454 | 0.961 | -0.307 | -1.083 |
| 15 | -0.889 | -0.805 | -0.727 | -0.727 | 0.988 | 0.656 | -0.727 | 0.947 | -0.259 | -0.049 | 0.025 | -0.981 | -1.844 | 0.216 | 0.025 | -0.049 |
| 16 | 0.245 | -0.213 | 0.025 | -0.087 | 0.578 | -0.169 | -0.588 | 0.326 | -0.259 | 0.025 | -0.466 | -0.525 | -0.259 | -0.41 | 0.796 | -0.307 |
| 17 | -0.805 | -0.525 | 0.093 | 0.904 | -0.805 | -0.087 | -1.638 | 0.578 | -0.727 | -0.41 | -1.195 | 1.09 | -2.103 | 0.187 | -0.41 | 0.494 |