Model info
| Transcription factor | SOX4 (GeneCards) | ||||||||
| Model | SOX4_HUMAN.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 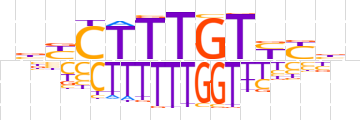 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 13 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bbYCTTTGTYYbb | ||||||||
| Best auROC (human) | 0.67 | ||||||||
| Best auROC (mouse) | 0.976 | ||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | 7 | ||||||||
| Aligned words | 350 | ||||||||
| TF family | SOX-related factors {4.1.1} | ||||||||
| TF subfamily | Group C {4.1.1.3} | ||||||||
| HGNC | HGNC:11200 | ||||||||
| EntrezGene | GeneID:6659 (SSTAR profile) | ||||||||
| UniProt ID | SOX4_HUMAN | ||||||||
| UniProt AC | Q06945 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | SOX4 expression | ||||||||
| ReMap ChIP-seq dataset list | SOX4 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 6.0 | 9.0 | 15.0 | 6.0 | 8.0 | 30.0 | 24.0 | 31.0 | 3.0 | 52.0 | 37.0 | 21.0 | 0.0 | 33.0 | 24.0 | 51.0 |
| 02 | 0.0 | 7.0 | 9.0 | 1.0 | 0.0 | 73.0 | 21.0 | 30.0 | 1.0 | 50.0 | 22.0 | 27.0 | 0.0 | 56.0 | 11.0 | 42.0 |
| 03 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 149.0 | 0.0 | 37.0 | 2.0 | 56.0 | 0.0 | 5.0 | 1.0 | 86.0 | 0.0 | 13.0 |
| 04 | 0.0 | 0.0 | 0.0 | 3.0 | 28.0 | 4.0 | 3.0 | 257.0 | 0.0 | 0.0 | 0.0 | 0.0 | 13.0 | 2.0 | 1.0 | 39.0 |
| 05 | 1.0 | 2.0 | 1.0 | 37.0 | 0.0 | 0.0 | 0.0 | 6.0 | 0.0 | 1.0 | 3.0 | 0.0 | 1.0 | 7.0 | 13.0 | 278.0 |
| 06 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 2.0 | 6.0 | 0.0 | 5.0 | 0.0 | 12.0 | 5.0 | 1.0 | 1.0 | 314.0 |
| 07 | 0.0 | 2.0 | 4.0 | 0.0 | 0.0 | 1.0 | 6.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 29.0 | 302.0 | 2.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 4.0 | 1.0 | 23.0 | 0.0 | 3.0 | 8.0 | 305.0 | 0.0 | 0.0 | 1.0 | 1.0 |
| 09 | 0.0 | 2.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 4.0 | 0.0 | 1.0 | 2.0 | 7.0 | 2.0 | 106.0 | 37.0 | 184.0 |
| 10 | 0.0 | 2.0 | 0.0 | 1.0 | 5.0 | 70.0 | 2.0 | 33.0 | 1.0 | 23.0 | 8.0 | 9.0 | 3.0 | 138.0 | 26.0 | 29.0 |
| 11 | 0.0 | 2.0 | 4.0 | 3.0 | 19.0 | 90.0 | 25.0 | 99.0 | 1.0 | 17.0 | 7.0 | 11.0 | 7.0 | 20.0 | 32.0 | 13.0 |
| 12 | 3.0 | 10.0 | 10.0 | 4.0 | 19.0 | 51.0 | 25.0 | 34.0 | 12.0 | 23.0 | 23.0 | 10.0 | 8.0 | 56.0 | 35.0 | 27.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.251 | -0.865 | -0.37 | -1.251 | -0.978 | 0.311 | 0.091 | 0.344 | -1.888 | 0.856 | 0.519 | -0.04 | -4.107 | 0.406 | 0.091 | 0.837 |
| 02 | -4.107 | -1.105 | -0.865 | -2.79 | -4.107 | 1.194 | -0.04 | 0.311 | -2.79 | 0.817 | 0.006 | 0.207 | -4.107 | 0.93 | -0.671 | 0.644 |
| 03 | -4.107 | -2.79 | -4.107 | -4.107 | -4.107 | 1.904 | -4.107 | 0.519 | -2.241 | 0.93 | -4.107 | -1.422 | -2.79 | 1.357 | -4.107 | -0.509 |
| 04 | -4.107 | -4.107 | -4.107 | -1.888 | 0.243 | -1.628 | -1.888 | 2.449 | -4.107 | -4.107 | -4.107 | -4.107 | -0.509 | -2.241 | -2.79 | 0.571 |
| 05 | -2.79 | -2.241 | -2.79 | 0.519 | -4.107 | -4.107 | -4.107 | -1.251 | -4.107 | -2.79 | -1.888 | -4.107 | -2.79 | -1.105 | -0.509 | 2.527 |
| 06 | -4.107 | -4.107 | -2.79 | -2.79 | -2.79 | -2.79 | -2.241 | -1.251 | -4.107 | -1.422 | -4.107 | -0.587 | -1.422 | -2.79 | -2.79 | 2.649 |
| 07 | -4.107 | -2.241 | -1.628 | -4.107 | -4.107 | -2.79 | -1.251 | -4.107 | -4.107 | -4.107 | -1.628 | -4.107 | -4.107 | 0.278 | 2.61 | -2.241 |
| 08 | -4.107 | -4.107 | -4.107 | -4.107 | -1.628 | -1.628 | -2.79 | 0.049 | -4.107 | -1.888 | -0.978 | 2.62 | -4.107 | -4.107 | -2.79 | -2.79 |
| 09 | -4.107 | -2.241 | -2.79 | -2.79 | -2.79 | -2.79 | -2.79 | -1.628 | -4.107 | -2.79 | -2.241 | -1.105 | -2.241 | 1.565 | 0.519 | 2.115 |
| 10 | -4.107 | -2.241 | -4.107 | -2.79 | -1.422 | 1.152 | -2.241 | 0.406 | -2.79 | 0.049 | -0.978 | -0.865 | -1.888 | 1.828 | 0.17 | 0.278 |
| 11 | -4.107 | -2.241 | -1.628 | -1.888 | -0.138 | 1.402 | 0.131 | 1.497 | -2.79 | -0.247 | -1.105 | -0.671 | -1.105 | -0.088 | 0.375 | -0.509 |
| 12 | -1.888 | -0.763 | -0.763 | -1.628 | -0.138 | 0.837 | 0.131 | 0.435 | -0.587 | 0.049 | 0.049 | -0.763 | -0.978 | 0.93 | 0.464 | 0.207 |